Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages - PubMed (original) (raw)
Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages
Jeroen Maertzdorf et al. J Clin Microbiol. 2004 Mar.
Abstract
The discovery of human metapneumovirus and its implications for respiratory tract disease have emphasized the need for a sensitive, specific, and rapid assay to detect this virus in a clinical setting. It recently became clear that human metapneumovirus can be grouped into at least four genetic lineages. Previously described assays for the detection of human metapneumovirus were developed by using limited sequence information and failed to detect viruses from all four genetic lineages with comparable sensitivities. Here we describe the development and evaluation of a real-time reverse transcriptase PCR assay that detects human metapneumovirus from the four known genetic lineages with equal specificities and sensitivities.
Figures
FIG. 1.
Hybridization blot showing detection of serial dilutions of viral RNAs of the prototype hMPV strains from genetic lineages A1 (NL/1/00), A2 (NL/17/00), B1 (NL/1/99), and B2 (NL/1/94) by using the conventional end-point RT-PCR assay targeting the hMPV L gene (15). Values above the lanes indicate the amounts of virus (in TCID50) from which RNA used in each reaction was obtained.
FIG. 2.
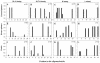
Entropy plots of oligonucleotide-annealing sites in hMPV sequences available from the GenBank database. The sequences recognized by the individual oligonucleotides were compared to all available hMPV sequences, and their heterogeneities are displayed as entropy values on the x axis. A higher entropy value indicates mismatches at a particular position in the oligonucleotide with a larger number of target sequences analyzed. The number of sequences upon which each plot was based is given in the upper right corner of each plot. Oligonucleotide positions are given in the 5′-3′ direction, with position 1 being the extreme 5′ nucleotide.
FIG. 3.
Alignment of sequences of primers and probes for the NL-N and ALT-N assays with the target sequences of the four prototype hMPV strains. Residue Y in the third position of the NL-N probe represents either a C or a T.
FIG. 4.
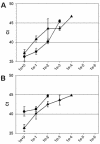
Real-Time RT-PCR amplification in the N (A) and L (B) gene assays of serially diluted RNAs of prototype hMPV strains. The values on the x axes indicate the amounts of RNA per reaction, ranging from undiluted (1e+0) to millionfold diluted (1e−6) RNAs. Ct values on the y axes indicate the first PCR cycle at which a positive signal was detected for each sample. Black circles and triangles represent the lineage A1 (NL/1/00) and A2 (NL/17/00) prototype virus strains, respectively. No amplification of viral RNA from lineage B viruses was detected in these experiments. Error bars indicate standard deviations for triplicate samples.
FIG. 5.
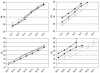
Real-time RT-PCR amplification of serially diluted RNAs from virus stocks (top panels) and RNA runoff transcripts from the nucleoprotein gene (bottom panels) of four prototype hMPV strains inthe NL-N assay (left panels) and the ALT-N assay (right panels). Black symbols are the same as those defined in the legend to Fig. 4. Grey circles and triangles represent the lineage B1 (NL/1/99) and B2 (NL/1/94) prototype virus strains, respectively. The values on the x axes in the top panels indicate the serial dilutions of RNA per reaction, ranging from 0-fold (1e−) to millionfold (1e−) dilutions. The values on the x axes in the bottom panels indicate the numbers of RNA molecules per reaction, ranging from 50 million (5e+7) to 50 (5e+1). Error bars indicate standard deviations. Ct, threshold cycle.
Similar articles
- Real-time reverse transcriptase PCR assay for improved detection of human metapneumovirus.
Klemenc J, Asad Ali S, Johnson M, Tollefson SJ, Talbot HK, Hartert TV, Edwards KM, Williams JV. Klemenc J, et al. J Clin Virol. 2012 Aug;54(4):371-5. doi: 10.1016/j.jcv.2012.05.005. Epub 2012 Jun 6. J Clin Virol. 2012. PMID: 22677006 Free PMC article. - Development of real-time RT-PCR for detection of human metapneumovirus and genetic analysis of circulating strains (2009-2011) in Pune, India.
Choudhary ML, Anand SP, Sonawane NS, Chadha MS. Choudhary ML, et al. Arch Virol. 2014 Feb;159(2):217-25. doi: 10.1007/s00705-013-1812-6. Epub 2013 Aug 9. Arch Virol. 2014. PMID: 23929232 Free PMC article. - Detection and characterisation of human metapneumovirus from children with acute respiratory symptoms in north-west England, UK.
Hopkins MJ, Redmond C, Shaw JM, Hart IJ, Hart CA, Smyth RL, Semple MG. Hopkins MJ, et al. J Clin Virol. 2008 Jul;42(3):273-9. doi: 10.1016/j.jcv.2008.03.011. Epub 2008 May 2. J Clin Virol. 2008. PMID: 18455957 - Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations.
Samransamruajkit R, Thanasugarn W, Prapphal N, Theamboonlers A, Poovorawan Y. Samransamruajkit R, et al. J Infect. 2006 Apr;52(4):254-63. doi: 10.1016/j.jinf.2005.07.001. Epub 2005 Sep 23. J Infect. 2006. PMID: 16183133 - Human metapneumovirus: a newly emerging respiratory pathogen.
Kahn JS. Kahn JS. Curr Opin Infect Dis. 2003 Jun;16(3):255-8. doi: 10.1097/00001432-200306000-00012. Curr Opin Infect Dis. 2003. PMID: 12821817 Review.
Cited by
- Co-circulation of four human coronaviruses (HCoVs) in Queensland children with acute respiratory tract illnesses in 2004.
Mackay IM, Arden KE, Speicher DJ, O'Neil NT, McErlean PK, Greer RM, Nissen MD, Sloots TP. Mackay IM, et al. Viruses. 2012 Apr;4(4):637-53. doi: 10.3390/v4040637. Epub 2012 Apr 23. Viruses. 2012. PMID: 22590689 Free PMC article. - Detection of novel polyomaviruses, TSPyV, HPyV6, HPyV7, HPyV9 and MWPyV in feces, urine, blood, respiratory swabs and cerebrospinal fluid.
Rockett RJ, Sloots TP, Bowes S, O'Neill N, Ye S, Robson J, Whiley DM, Lambert SB, Wang D, Nissen MD, Bialasiewicz S. Rockett RJ, et al. PLoS One. 2013 May 8;8(5):e62764. doi: 10.1371/journal.pone.0062764. Print 2013. PLoS One. 2013. PMID: 23667518 Free PMC article. - Identification of new pathogens in the intraocular fluid of patients with uveitis.
de Groot-Mijnes JD, de Visser L, Zuurveen S, Martinus RA, Völker R, ten Dam-van Loon NH, de Boer JH, Postma G, de Groot RJ, van Loon AM, Rothova A. de Groot-Mijnes JD, et al. Am J Ophthalmol. 2010 Nov;150(5):628-36. doi: 10.1016/j.ajo.2010.05.015. Epub 2010 Aug 5. Am J Ophthalmol. 2010. PMID: 20691420 Free PMC article. - Comparative Viral Sampling in the Sinonasal Passages; Different Viruses at Different Sites.
Goggin RK, Bennett CA, Bassiouni A, Bialasiewicz S, Vreugde S, Wormald PJ, Psaltis AJ. Goggin RK, et al. Front Cell Infect Microbiol. 2018 Sep 19;8:334. doi: 10.3389/fcimb.2018.00334. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30283747 Free PMC article. - A multiplex one-tube nested real time RT-PCR assay for simultaneous detection of respiratory syncytial virus, human rhinovirus and human metapneumovirus.
Feng ZS, Zhao L, Wang J, Qiu FZ, Zhao MC, Wang L, Duan SX, Zhang RQ, Chen C, Qi JJ, Fan T, Li GX, Ma XJ. Feng ZS, et al. Virol J. 2018 Oct 30;15(1):167. doi: 10.1186/s12985-018-1061-0. Virol J. 2018. PMID: 30376870 Free PMC article.
References
- Boivin, G., Y. Abed, G. Pelletier, L. Ruel, D. Moisan, S. Cote, T. C. Peret, D. D. Erdman, and L. J. Anderson. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330-1334. - PubMed
- Esper, F., D. Boucher, C. Weibel, R. A. Martinello, and J. S. Kahn. 2003. Human metapneumovirus infection in the Unites States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407-1410. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources