Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae - PubMed (original) (raw)
Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae
Fulvio Reggiori et al. Mol Biol Cell. 2004 May.
Abstract
The Cvt pathway is a biosynthetic transport route for a distinct subset of resident yeast vacuolar hydrolases, whereas macroautophagy is a nonspecific degradative mechanism that allows cell survival during starvation. Yet, these two vacuolar trafficking pathways share a number of identical molecular components and are morphologically very similar. For example, one of the hallmarks of both pathways is the formation of double-membrane cytosolic vesicles that sequester cargo before vacuolar delivery. The origin of the vesicle membrane has been controversial and various lines of evidence have implicated essentially all compartments of the endomembrane system. Despite the analogies between the Cvt pathway and autophagy, earlier work has suggested that the origin of the engulfing vesicle membranes is different; the endoplasmic reticulum is proposed to be required only for autophagy. In contrast, in this study we demonstrate that the endoplasmic reticulum and/or Golgi complex, but not endosomal compartments, play an important role for both yeast transport routes. Along these lines, we demonstrate that Berkeley bodies, a structure generated from the Golgi complex in sec7 cells, are immunolabeled with Atg8, a structural component of autophagosomes. Finally, we also show that none of the yeast t-SNAREs are located at the preautophagosomal structure, the presumed site of double-membrane vesicle formation. Based on our results, we propose two models for Cvt vesicle biogenesis.
Figures
Figure 1.
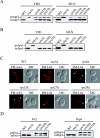
The late endosome does not participate in the biogenesis of the PAS. (A) Wild-type (WT), _atg1_Δ, vps4Δ, _vps23_Δ, _vps27_Δ, and _vps28_Δ, and (B) WT, _atg1_Δ, vps5Δ, _vps29_Δ, and _vps35_Δ cells in the BY4742 background grown in YPD or nitrogen starved in SD-N medium for 4 h were TCA precipitated. Acetone washed proteins were then resolved by SDS-PAGE and prApe1 maturation analyzed by immunoblot with serum to Ape1. All strains showed normal prApe1 maturation by either the Cvt pathway (YPD) or autophagy (SD-N). (C) Morphological anomalies of class E vps mutants. Wild-type (WT), _atg1_Δ, vps4Δ, _vps23_Δ, _vps27_Δ, and _vps28_Δ cells were labeled with FM 4–64 and imaged as described previously (Reggiori et al., 2003). Arrowheads indicate enlarged endosomes. All four class E vps mutants accumulated an aberrant endosomal structure. DIC, differential interference contrast. (D) Vacuolar protease missorting in retromer mutants. Cell extracts analyzed in B were probed with anti-Prc1 and anti-Pep4 antisera. Retromer deletants accumulated unprocessed Prc1 (p2Prc1) and Pep4 (prPep4).
Figure 2.
The VFT complex, Tlg2, and all other t-SNAREs do not localize to the PAS. (A) The VFT complex does not localize to the PAS. Wild-type (SEY6210) cells expressing either Vps51-YFP (PSY82) or Vps54-YFP (PSY83) were transformed with the pRS316ECFP-Aut7 plasmid. The resulting strains were grown in SMD medium to an early log stage and analyzed with a fluorescence microscope. (B) Tlg1 and Tlg2 do not localize to the PAS. The wild-type (SEY6210) strain was cotransformed with plasmids encoding either YFP-Tlg1 or YFP-Tlg2 and CFP-Atg8. Transformed cells were analyzed as in A. (C) Tlg1 colocalizes with Tlg2 and partially with Snc1. The wild-type (SEY6210) strain was cotransformed with plasmids encoding YFP-Tlg1 and either CFP-Tlg2 or CFP-Snc1. Transformed cells were analyzed as in A. (D) Tlg1 and Tlg2 do not fractionate with the PAS. The wild-type (HAY456) strain expressing N-terminal PA-tagged Atg9 was transformed with the plasmid expressing YFP-Tlg2. Spheroplasts from the transformed cells were grown in rich medium until mid-log phase, osmotically lysed, and the membranes fractionated on a sucrose density gradient as described in MATERIALS AND METHODS. In total, 14 fractions were collected from the top of the gradient, resolved by SDS-PAGE, and membranes were probed with antisera to PA, Atg8, GFP, and Tlg1. (E) None of the yeast t-SNAREs localize to the PAS. The same gradient fractions analyzed in D were also probed for Vam3, Sed5, Sso2, and Pep12. None of these t-SNAREs colocalized with the PAS marker Atg9.
Figure 3.
The PAS and early endosomes are different structures. (A) Snc1 does not localize to the PAS. Wild-type (SEY6210) cells carrying plasmids expressing both YFP-Snc1 and CFP-Atg8 were grown in SMD medium to an early log stage and analyzed with a fluorescence microscope. (B) Snc1 recycling is blocked in the _rcy1_Δ strain. The wild-type (WT) and _rcy1_Δ cells in the BY4742 background were transformed with a plasmid expressing the GFP-Snc1 chimera. Transformed cells were grown to mid-log stage in SMD medium and imaged with a fluorescence microscope. In wild-type cells, Snc1 was concentrated on the plasma membrane, whereas it was accumulated in internal structures in the mutant. (C) Precursor Ape1 processing is normal in the _rcy1_Δ strain. Wild-type (WT), _atg9_Δ and _rcy1_Δ cells in the BY4742 background grown either in YPD or nitrogen starved in SD-N medium for 4 h were TCA precipitated. Acetone washed proteins were then resolved by SDS-PAGE and prApe1 maturation was analyzed by immunoblot with serum to Ape1. Both the wild-type and _rcy1_Δ strains showed normal prApe1 maturation either when grown in YPD medium or nitrogen starved for 4 h.
Figure 4.
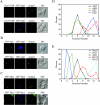
Both PtdIns 3-kinase complexes generate the PtdIns(3)P on the early endosome surface. (A) Wild-type (WT), _vps34_Δ, _vps38_Δ, _atg14_Δ, and _atg14_Δ _vps38_Δ cells in the BY4742 background were transformed with a plasmid expressing either GFP-Ape1 or GFP-Snc1. Transformed cells were grown to a mid-log stage in SMD medium and imaged with a fluorescence microscope. GFP-Ape1 was delivered to the vacuole lumen in wild-type and _vps38_Δ cells but not in _vps34_Δ, _atg14_Δ and _atg14_Δ _vps38_Δ mutants where it was accumulated at the PAS. GFP-Snc1 displayed normal cycling in wild-type, _vps38_Δ, and _atg14_Δ cells but not in _vps34_Δ and _atg14_Δ _vps38_Δ mutants, indicating that its transport between the early endosome and Golgi complex depends on the PtdIns(3)P produced by both PtdIns 3-kinase complexes. (B) The maturation of endogenous prApe1 is blocked in _vps34_Δ, _atg14_Δ, and _atg14_Δ _vps38_Δ strains.
Figure 5.
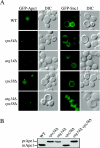
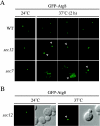
Early secretory mutants accumulate an aberrant PAS. (A) The wild-type (WT, SEY6210), sec12 (FBY217), and sec7 (AFM69–1A) strains were transformed with a plasmid expressing GFP-Atg8 under the control of the CUP1 promoter. Transformed cells were grown at 24°C to early log stage in SMD medium and imaged with a fluorescence microscope. Cultures were then transferred to 37°C, and cells were imaged every 30 min. Between 90 min and 2 h at restrictive temperature, sec12 and sec7 cells started to display the presence of aberrant PAS some of which assumed toroid and ellipsoid forms, suggesting that early secretory mutations affect proper PAS organization. Examples of images taken after 2 h are shown and toroid structures indicated with a white arrowhead. (B) Overexpression of Atg8 does not promote the formation of aberrant PAS structures in early secretory mutants. The sec12 strain (FBY217) was transformed with a plasmid carrying a GFP-Atg8 fusion driven by the authentic ATG8 promoter. Transformed cells were analyzed as in A. GFP-Atg8–marked toroid and ellipsoid structures were observed in sec12 cells incubated at 37°C for 2 h.
Figure 6.
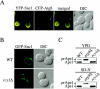
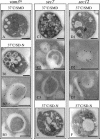
Atg8 is targeted to the Berkeley bodies in sec7 and sec12 cells. The vam3ts (A and B), s_ec7_ (C and E), and sec12 (D and F) strains expressing a triple hemagglutinin-ATG8 fusion under the control of the authentic ATG8 promoter were grown at 24°C in YPD medium and then shifted to 37°C for 3 h in either the same medium (A, C, and D) or in SD-N (B, E, and F). Cells were collected before and after the temperature and the media shifts. All samples were prepared for immunoelectron microscopy as described in MATERIALS AND METHODS. Black bar, 1 μm; white bar, 0.2 μm.
Figure 7.
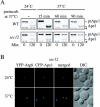
A functional Golgi complex is essential for autophagy. (A) The sec12 (FBY217) and sec7 (AFM69-1A) strains were transformed with both a multi-copy plasmid expressing Ape1 and either an empty vector (pRS415) or one carrying the wild-type allele of the mutated gene (pSEC12 or pSEC7, respectively). Transformed cells and two wild-type strains (WT, SEY6210, and BY4742) were grown to 1 OD600 in SMD medium at 24°C. After washing twice with water, cells were nitrogen starved in SD-N medium for 4 h at 37°C before collecting the proteins by TCA precipitation. Polypeptides were finally resolved by SDS-PAGE and prApe1 maturation analyzed by immunoblot with serum to Ape1. (B) The sec12 (FRY208) and sec7 (FRY209) strains expressing Pho8Δ60 and carrying either an empty vector (pRS415) or a plasmid complementing the corresponding sec mutation (pSEC12 or pSEC7, respectively) were shifted from SMD medium at 24°C (black bars) to SD-N medium at 34°C (white bars) for 4 h. Autophagy induction was determined by a Pho8 activity assay. Results were expressed as percentage of the activity measured for the complemented strains starved for nitrogen.
Figure 8.
Early stages of the secretory pathway are essential for the Cvt pathway. (A) Precursor Ape1 transport is affected in sec12 cells. Wild-type (WT and SEY6210) and sec12 (FBY217) cells were grown at 24°C and then shifted to 37°C for 15, 60, and 90 min, respectively. Both strains were pulse-labeled and subjected to a nonradioactive chase as described in MATERIALS AND METHODS. Samples were removed at the beginning (0 min) and at the end (120 min) of the chase period. Proteins were TCA precipitated and then immunoprecipitated with antiserum to Ape1. Precursor Ape1 transport was perturbed in sec12 cells at prolonged restrictive temperatures but not in wild-type cells. The asterisk marks a cross-reacting band. (B) Precursor Ape1 is not correctly targeted to the PAS when ER exit is blocked. The sec12 strain (FBY217) was transformed with plasmids expressing CFP-Ape1 and YFP-Atg8. Transformed cells were grown at 24°C in SMD and imaged. The temperature was then shifted to 37°C and cells imaged again after 2 h. CFP-Ape1 and YFP-Atg8 colocalized to the PAS when the cells were grown at 24°C. Incubation at restrictive temperature for 2 h disrupted the correct CFP-Ape1 recruitment to the PAS marked with YFP-Atg8 in 50–60% of the imaged cells.
Figure 9.
Models for the PAS and double-membrane vesicle biogenesis. In the maturation model, integral membrane proteins involved in Cvt vesicle and autophagosome formation are segregated in a compartment derived from the membrane source. This organelle then matures into the PAS and finally becomes the double-membrane vesicle in a process balanced by the retrieval of specific proteins back to the donor compartment. In the assembly model, several sets of lipid bilayers are derived from the membrane source by a maturation process similar to the one described above. Those structures are then assembled into double-membrane vesicles at the PAS. Both models suggest a SNARE-independent fusion event required for the sealing of double-membrane vesicles and a retrieval transport route (dashed lines). During Cvt vesicle formation, this route involves sorting nexins and possibly Tlg1, Tlg2, Vps45, and the VFT complex.
Similar articles
- Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway.
Meiling-Wesse K, Epple UD, Krick R, Barth H, Appelles A, Voss C, Eskelinen EL, Thumm M. Meiling-Wesse K, et al. J Biol Chem. 2005 Sep 30;280(39):33669-78. doi: 10.1074/jbc.M501701200. Epub 2005 Aug 3. J Biol Chem. 2005. PMID: 16079147 - TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy.
Shirahama-Noda K, Kira S, Yoshimori T, Noda T. Shirahama-Noda K, et al. J Cell Sci. 2013 Nov 1;126(Pt 21):4963-73. doi: 10.1242/jcs.131318. Epub 2013 Aug 28. J Cell Sci. 2013. PMID: 23986483 - Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion.
Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Ishihara N, et al. Mol Biol Cell. 2001 Nov;12(11):3690-702. doi: 10.1091/mbc.12.11.3690. Mol Biol Cell. 2001. PMID: 11694599 Free PMC article. - SNAREs and NSF in targeted membrane fusion.
Hay JC, Scheller RH. Hay JC, et al. Curr Opin Cell Biol. 1997 Aug;9(4):505-12. doi: 10.1016/s0955-0674(97)80026-9. Curr Opin Cell Biol. 1997. PMID: 9261050 Review. - An Overview of Protein Secretion in Yeast and Animal Cells.
Guo Y, Yang F, Tang X. Guo Y, et al. Methods Mol Biol. 2017;1662:1-17. doi: 10.1007/978-1-4939-7262-3_1. Methods Mol Biol. 2017. PMID: 28861813 Review.
Cited by
- Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae.
Nazarko TY, Huang J, Nicaud JM, Klionsky DJ, Sibirny AA. Nazarko TY, et al. Autophagy. 2005 Apr;1(1):37-45. doi: 10.4161/auto.1.1.1512. Epub 2005 Apr 30. Autophagy. 2005. PMID: 16874038 Free PMC article. - An overview of the molecular mechanism of autophagy.
Yang Z, Klionsky DJ. Yang Z, et al. Curr Top Microbiol Immunol. 2009;335:1-32. doi: 10.1007/978-3-642-00302-8_1. Curr Top Microbiol Immunol. 2009. PMID: 19802558 Free PMC article. Review. - The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation.
Müller M, Schmidt O, Angelova M, Faserl K, Weys S, Kremser L, Pfaffenwimmer T, Dalik T, Kraft C, Trajanoski Z, Lindner H, Teis D. Müller M, et al. Elife. 2015 Apr 22;4:e07736. doi: 10.7554/eLife.07736. Elife. 2015. PMID: 25902403 Free PMC article. - Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles.
Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Karanasios E, et al. Nat Commun. 2016 Aug 11;7:12420. doi: 10.1038/ncomms12420. Nat Commun. 2016. PMID: 27510922 Free PMC article. - Unconventional secretion of Acb1 is mediated by autophagosomes.
Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Duran JM, et al. J Cell Biol. 2010 Feb 22;188(4):527-36. doi: 10.1083/jcb.200911154. Epub 2010 Feb 15. J Cell Biol. 2010. PMID: 20156967 Free PMC article.
References
- Abeliovich, H., Grote, E., Novick, P., and Ferro-Novick, S. (1998). Tlg2p, a yeast syntaxin homolog that resides on the Golgi and endocytic structures. J. Biol. Chem. 273, 11719–11727. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases