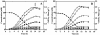Engineering Lactococcus lactis for production of mannitol: high yields from food-grade strains deficient in lactate dehydrogenase and the mannitol transport system - PubMed (original) (raw)
Engineering Lactococcus lactis for production of mannitol: high yields from food-grade strains deficient in lactate dehydrogenase and the mannitol transport system
Paula Gaspar et al. Appl Environ Microbiol. 2004 Mar.
Abstract
Mannitol is a sugar polyol claimed to have health-promoting properties. A mannitol-producing strain of Lactococcus lactis was obtained by disruption of two genes of the phosphoenolpyruvate (PEP)-mannitol phosphotransferase system (PTS(Mtl)). Genes mtlA and mtlF were independently deleted by double-crossover recombination in strain L. lactis FI9630 (a food-grade lactate dehydrogenase-deficient strain derived from MG1363), yielding two mutant (Delta ldh Delta mtlA and Delta ldh Delta mtlF) strains. The new strains, FI10091 and FI10089, respectively, do not possess any selection marker and are suitable for use in the food industry. The metabolism of glucose in nongrowing cell suspensions of the mutant strains was characterized by in vivo (13)C-nuclear magnetic resonance. The intermediate metabolite, mannitol-1-phosphate, accumulated intracellularly to high levels (up to 76 mM). Mannitol was a major end product, one-third of glucose being converted to this hexitol. The double mutants, in contrast to the parent strain, were unable to utilize mannitol even after glucose depletion, showing that mannitol was taken up exclusively by PEP-PTS(Mtl). Disruption of this system completely blocked mannitol transport in L. lactis, as intended. In addition to mannitol, approximately equimolar amounts of ethanol, 2,3-butanediol, and lactate were produced. A mixed-acid fermentation (formate, ethanol, and acetate) was also observed during growth under controlled conditions of pH and temperature, but mannitol production was low. The reasons for the alteration in the pattern of end products under nongrowing and growing conditions are discussed, and strategies to improve mannitol production during growth are proposed.
Figures
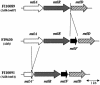
FIG. 1.
Schematic representation of the mannitol operon in the chromosome of the parent (FI9630) and mutant strains obtained by gene replacement. The deletions were confirmed by PCR analysis. _mtlA_′, truncated mtlA.
FIG. 2.
Growth, glucose consumption, and end product formation by FI10089 (A) and FI9630 (B). •, glucose; □, formate; ▵, ethanol; ▪, acetate; ▿, acetoin; ▾, 2,3-butanediol; ⧫, lactate; ○, mannitol; x, biomass. At the time indicated by the arrow, a culture sample was withdrawn for ethanol extraction and quantification of intracellular mannitol. Data are from a representative experiment in which the error for each data point is ≤15%.
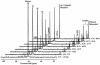
FIG. 3.
Sequence of 13C-NMR spectra acquired during the metabolism of 40 mM [6-13C]glucose by a cell suspension of FI10089 under anaerobic conditions at 30°C. Cells were suspended in 50 mM KPi (pH 6.5) at a concentration of 15 mg of protein ml−1. The pH was kept constant by automatic addition of NaOH. Each spectrum represents 30 s of acquisition. Glucose was added at time zero, and each spectrum was acquired during the indicated interval and processed with a 12-Hz line broadening.
FIG. 4.
Kinetics of glucose consumption by nongrowing cells of L. lactis FI10089 as determined in vivo by 13C-NMR. (A) Pools of intracellular metabolites. (B) Kinetics of glucose consumption and end product formation. Data are from an individual experiment, except for those representing mannitol and mannitol-1-phosphate pools, which were estimated by combining data from parallel 13C-NMR experiments with a supply of [6-13C]glucose or [1-13C]glucose, as described in the Results section. The error varied from 2% (end products) to 15% in the case of intracellular metabolites. •, glucose; ▾, 2,3-butanediol; ▵, ethanol; ⧫, lactate; ▿, acetoin; ▪, acetate; ○, mannitol; ◊, Mtl1P; ▴, FBP; ★, 3-PGA; +, PEP.
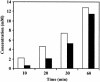
FIG. 5.
Total and extracellular mannitol during glucose metabolism in nongrowing cells of L. lactis FI10089 as determined by 13C-NMR. For the calculations, the total mannitol was assumed as extracellular. □, total mannitol; ▪, extracellular mannitol.
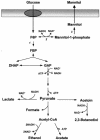
FIG. 6.
Pathways proposed for glucose metabolism and mannitol synthesis in L. lactis FI10089 carrying a double deletion of the genes ldh and mtlF. 1, phosphofructokinase; 2, mannitol-1-phosphate dehydrogenase; 3, mannitol-1-phosphatase.
Similar articles
- Metabolic characterization of Lactococcus lactis deficient in lactate dehydrogenase using in vivo 13C-NMR.
Neves AR, Ramos A, Shearman C, Gasson MJ, Almeida JS, Santos H. Neves AR, et al. Eur J Biochem. 2000 Jun;267(12):3859-68. doi: 10.1046/j.1432-1327.2000.01424.x. Eur J Biochem. 2000. PMID: 10849005 - High yields of 2,3-butanediol and mannitol in Lactococcus lactis through engineering of NAD⁺ cofactor recycling.
Gaspar P, Neves AR, Gasson MJ, Shearman CA, Santos H. Gaspar P, et al. Appl Environ Microbiol. 2011 Oct;77(19):6826-35. doi: 10.1128/AEM.05544-11. Epub 2011 Aug 12. Appl Environ Microbiol. 2011. PMID: 21841021 Free PMC article. - Metabolic engineering of mannitol production in Lactococcus lactis: influence of overexpression of mannitol 1-phosphate dehydrogenase in different genetic backgrounds.
Wisselink HW, Mars AE, van der Meer P, Eggink G, Hugenholtz J. Wisselink HW, et al. Appl Environ Microbiol. 2004 Jul;70(7):4286-92. doi: 10.1128/AEM.70.7.4286-4292.2004. Appl Environ Microbiol. 2004. PMID: 15240312 Free PMC article. - Metabolic engineering of Lactococcus lactis: the impact of genomics and metabolic modelling.
Kleerebezem M, Boels IC, Groot MN, Mierau I, Sybesma W, Hugenholtz J. Kleerebezem M, et al. J Biotechnol. 2002 Sep 25;98(2-3):199-213. doi: 10.1016/s0168-1656(02)00132-3. J Biotechnol. 2002. PMID: 12141987 Review. - [Recent advances in the bioproduction of mannitol].
Yan D, Cai X, Xue H, Zhen N, Wu Y, Liu Z, Li M, Zheng Y. Yan D, et al. Sheng Wu Gong Cheng Xue Bao. 2024 Aug 25;40(8):2626-2643. doi: 10.13345/j.cjb.240220. Sheng Wu Gong Cheng Xue Bao. 2024. PMID: 39174473 Review. Chinese.
Cited by
- Deciphering the Regulation of the Mannitol Operon Paves the Way for Efficient Production of Mannitol in Lactococcus lactis.
Xiao H, Bang-Berthelsen CH, Jensen PR, Solem C. Xiao H, et al. Appl Environ Microbiol. 2021 Jul 27;87(16):e0077921. doi: 10.1128/AEM.00779-21. Epub 2021 Jul 27. Appl Environ Microbiol. 2021. PMID: 34105983 Free PMC article. - High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering.
Ladero V, Ramos A, Wiersma A, Goffin P, Schanck A, Kleerebezem M, Hugenholtz J, Smid EJ, Hols P. Ladero V, et al. Appl Environ Microbiol. 2007 Mar;73(6):1864-72. doi: 10.1128/AEM.02304-06. Epub 2007 Jan 19. Appl Environ Microbiol. 2007. PMID: 17261519 Free PMC article. - System estimation from metabolic time-series data.
Goel G, Chou IC, Voit EO. Goel G, et al. Bioinformatics. 2008 Nov 1;24(21):2505-11. doi: 10.1093/bioinformatics/btn470. Epub 2008 Sep 4. Bioinformatics. 2008. PMID: 18772153 Free PMC article. - Physiological and Transcriptional Responses of Different Industrial Microbes at Near-Zero Specific Growth Rates.
Ercan O, Bisschops MM, Overkamp W, Jørgensen TR, Ram AF, Smid EJ, Pronk JT, Kuipers OP, Daran-Lapujade P, Kleerebezem M. Ercan O, et al. Appl Environ Microbiol. 2015 Sep 1;81(17):5662-70. doi: 10.1128/AEM.00944-15. Epub 2015 Jun 5. Appl Environ Microbiol. 2015. PMID: 26048933 Free PMC article. Review. - The cwp66 Gene Affects Cell Adhesion, Stress Tolerance, and Antibiotic Resistance in Clostridioides difficile.
Zhou Q, Rao F, Chen Z, Cheng Y, Zhang Q, Zhang J, Guan Z, He Y, Yu W, Cui G, Qi X, Hong W. Zhou Q, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0270421. doi: 10.1128/spectrum.02704-21. Epub 2022 Mar 31. Microbiol Spectr. 2022. PMID: 35357205 Free PMC article.
References
- Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous