Specificity of microRNA target selection in translational repression - PubMed (original) (raw)
. 2004 Mar 1;18(5):504-11.
doi: 10.1101/gad.1184404. Epub 2004 Mar 10.
Affiliations
- PMID: 15014042
- PMCID: PMC374233
- DOI: 10.1101/gad.1184404
Specificity of microRNA target selection in translational repression
John G Doench et al. Genes Dev. 2004.
Abstract
MicroRNAs (miRNAs) are a class of noncoding RNAs found in organisms as evolutionarily distant as plants and mammals, yet most of the mRNAs they regulate are unknown. Here we show that the ability of an miRNA to translationally repress a target mRNA is largely dictated by the free energy of binding of the first eight nucleotides in the 5' region of the miRNA. However, G:U wobble base-pairing in this region interferes with activity beyond that predicted on the basis of thermodynamic stability. Furthermore, an mRNA can be simultaneously repressed by more than one miRNA species. The level of repression achieved is dependent on both the amount of mRNA and the amount of available miRNA complexes. Thus, predicted miRNA:mRNA interactions must be viewed in the context of other potential interactions and cellular conditions.
Figures
Figure 1.
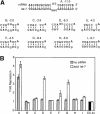
5′ region of the miRNA determines translational repression. (A) Schematic of the CXCR4 siRNA, antisense strand, base-pairing to a designed 3′-UTR-binding site. The two 3′-most nucleotides are deoxythymidines. Mutations were made in the mRNA to form mismatches with the siRNA. In each case, the 2-nt sequence of the mRNA was mutated to that of the siRNA. For example, mutant B contains a GU-to-CA mutation. (B) Luciferase assay of mutant constructs. Constructs were transfected ±siRNA, and fold repression was determined. The upper broken line corresponds to repression with four original sites, and the lower broken line corresponds to repression with two original sites flanking two binding sites for an unrelated siRNA (targeting GFP), and thus serves as the lower bound for repression. The experiment was performed three times, and averages are presented ±standard deviation. (C) Ribonuclease protection assay of steady-state mRNA levels. The upper band corresponds to firefly luciferase mRNA (control), and the lower band to Renilla luciferase mRNA (targeted). Lane 12 is 5% of input probe, and lane 11 shows that no species are protected in untransfected HeLa cells. 4x is the construct with four original CXCR4 sites, and mutants A, G, and H are described in A. The Renilla mRNA level was normalized to the firefly, and then the fold change was calculated for each construct, dividing the +siRNA value into the –siRNA value; a value <1 indicates a decrease in relative Renilla mRNA levels. (D) Twelve additional mutants with alterations in the binding site for the first 8 nt of the miRNA along with mutants F, G, H, and I from A. The structure predicted by mFold is shown, and the original binding site is shown for comparison. The two numbers above each binding site correspond to the fold repression achieved and the calculated ΔG value. (E) ΔG for the first 8 nt of the miRNA binding to the mRNA, plotted against fold repression, for the mutants in D as well as mutants F–I from A. The broken lines correspond to the same bounds as in B.
Figure 2.
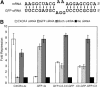
3′ region of the miRNA is rarely critical for repression. (A) Nine mutants with alterations in the binding site for the 3′ region of the miRNA. The structure predicted by mFold is shown, and the original binding site is shown for comparison. The nine sites shown are mutants A–E from Figure 1A, and four additional mutant constructs. The two numbers above each binding site correspond to the fold repression achieved and the calculated ΔG value. (B) ΔG of the 3′ region of the miRNA binding to the mRNA was calculated, and plotted against fold repression (±standard deviation from three independent experiments). The horizontal broken lines are the same as in Figure 1. (C) Effect of combined 5′- and 3′-binding site mutations. The left column shows the original binding site and two 5′-binding site mutant constructs. The number centered above the binding site is the fold repression achieved, and the smaller numbers are the ΔG values for the binding of the 5′ and 3′regions of the miRNA. Each construct on the left was then mutated in the 3′-region-binding site.
Figure 3.
G:U wobble in the 5′ region of the miRNA hinders repression. The 5′ region of the CXCR4 siRNA binding to the mRNA is shown, as well as four mutant constructs that create G:U wobble pairing. These constructs were assayed and plotted on top of the data presented in Figure 1E. Arrows point from the original binding site to the four mutant constructs, and are labeled with the position of the G:U wobble. Data points indicate the average of three independent experiments.
Figure 4.
Endogenous let-7a confirms importance of miRNA 5′ region. (A) Schematic of a 3′-UTR-binding site, and its predicted interaction with endogenous let-7a, along with eight mutant binding sites for the 5′ region of endogenous let-7a, together with the ΔG value. Constructs G, H, and I contain G:U wobble base pairs. (B) Fold repression for the various constructs shown in A. The fold repression achieved by endogenous let-7a is in gray. Expression values were first normalized internally to firefly luciferase expression, then across samples to the control construct, with four CXCR4 sites, shown in black. The constructs were then transfected with additional let-7a, and the fold repression is shown in white, again normalized to the expression of the control CXCR4–4x construct. Values are averages from three independent experiments, ±standard deviation.
Figure 5.
Distance requirements for miRNA accessibility. The binding sites inserted between two original CXCR4 sites are shown; for clarity, one of the CXCR4 siRNAs is shown in gray. The distance between the two sites was progressively reduced, until the 5′ region of one site moved into the 3′ region of the adjacent site. The fold repression achieved is indicated to the right of each schematic, the average of three independent experiments.
Figure 6.
Two miRNAs can simultaneously repress an mRNA. (A) Schematic of a binding site for an siRNA originally used to target GFP. (B) Four constructs were transfected with either the GFP siRNA, the CXCR4 siRNA, both siRNAs, or no siRNA. One construct had four CXCR4 sites, one had four GFP sites, and two constructs had two of each, in the arrangement indicated. Fold repression was determined, normalized to the no siRNA transfection. The average of three independent experiments is shown, ±standard deviation.
Comment in
- Great expectations of small RNAs.
Schuldt A. Schuldt A. Nat Rev Mol Cell Biol. 2010 Oct;11(10):676. doi: 10.1038/nrm2987. Nat Rev Mol Cell Biol. 2010. PMID: 20861869 No abstract available.
Similar articles
- Analysis of microRNA-target interactions by a target structure based hybridization model.
Long D, Chan CY, Ding Y. Long D, et al. Pac Symp Biocomput. 2008:64-74. Pac Symp Biocomput. 2008. PMID: 18232104 - Is the Efficiency of RNA Silencing Evolutionarily Regulated?
Ui-Tei K. Ui-Tei K. Int J Mol Sci. 2016 May 12;17(5):719. doi: 10.3390/ijms17050719. Int J Mol Sci. 2016. PMID: 27187367 Free PMC article. Review. - miRNA Targeting: Growing beyond the Seed.
Chipman LB, Pasquinelli AE. Chipman LB, et al. Trends Genet. 2019 Mar;35(3):215-222. doi: 10.1016/j.tig.2018.12.005. Epub 2019 Jan 9. Trends Genet. 2019. PMID: 30638669 Free PMC article. Review. - Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii.
Yamasaki T, Voshall A, Kim EJ, Moriyama E, Cerutti H, Ohama T. Yamasaki T, et al. Plant J. 2013 Dec;76(6):1045-56. doi: 10.1111/tpj.12354. Plant J. 2013. PMID: 24127635 - MicroRNA targeting specificity in mammals: determinants beyond seed pairing.
Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. Grimson A, et al. Mol Cell. 2007 Jul 6;27(1):91-105. doi: 10.1016/j.molcel.2007.06.017. Mol Cell. 2007. PMID: 17612493 Free PMC article.
Cited by
- A polymorphism in the 3'-untranslated region of the NPM1 gene causes illegitimate regulation by microRNA-337-5p and correlates with adverse outcome in acute myeloid leukemia.
Cheng CK, Kwan TK, Cheung CY, Ng K, Liang P, Cheng SH, Chan NP, Ip RK, Wong RS, Lee V, Li CK, Yip SF, Ng MH. Cheng CK, et al. Haematologica. 2013 Jun;98(6):913-7. doi: 10.3324/haematol.2012.073015. Epub 2012 Oct 12. Haematologica. 2013. PMID: 23065518 Free PMC article. - Identification and characterization of the miRNA transcriptome of Ovis aries.
Zhang S, Zhao F, Wei C, Sheng X, Ren H, Xu L, Lu J, Liu J, Zhang L, Du L. Zhang S, et al. PLoS One. 2013;8(3):e58905. doi: 10.1371/journal.pone.0058905. Epub 2013 Mar 13. PLoS One. 2013. PMID: 23516575 Free PMC article. - MicroRNA Targets PAP1 to Mediate Melanization in Plutella xylostella (Linnaeus) Infected by Metarhizium anisopliae.
Zhang Z, Jin F, Huang J, Mandal S, Zeng L, Zafar J, Xu X. Zhang Z, et al. Int J Mol Sci. 2024 Jan 17;25(2):1140. doi: 10.3390/ijms25021140. Int J Mol Sci. 2024. PMID: 38256210 Free PMC article. - Oscillating glucose induces microRNA-185 and impairs an efficient antioxidant response in human endothelial cells.
La Sala L, Cattaneo M, De Nigris V, Pujadas G, Testa R, Bonfigli AR, Genovese S, Ceriello A. La Sala L, et al. Cardiovasc Diabetol. 2016 Apr 30;15:71. doi: 10.1186/s12933-016-0390-9. Cardiovasc Diabetol. 2016. PMID: 27137793 Free PMC article. - Experimental procedures to identify and validate specific mRNA targets of miRNAs.
Elton TS, Yalowich JC. Elton TS, et al. EXCLI J. 2015 Jul 2;14:758-90. doi: 10.17179/excli2015-319. eCollection 2015. EXCLI J. 2015. PMID: 27047316 Free PMC article. Review.
References
- Abrahante J.E., Daul, A.L., Li, M., Volk, M.L., Tennessen, J.M., Miller, E.A., and Rougvie, A.E. 2003. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev. Cell 4: 625–637. - PubMed
- Bartel D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell 116: 281–297. - PubMed
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. - PubMed
- Brennecke J., Hipfner, D.R., Stark, A., Russell, R.B., and Cohen, S.M. 2003. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P01 CA042063/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- R37-GM34277/GM/NIGMS NIH HHS/United States
- R37 GM034277/GM/NIGMS NIH HHS/United States
- P01-CA42063/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials