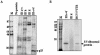Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem-loop structures of the 5'-untranslated region - PubMed (original) (raw)
Inhibition of hepatitis C virus IRES-mediated translation by small RNAs analogous to stem-loop structures of the 5'-untranslated region
Partho Sarothi Ray et al. Nucleic Acids Res. 2004.
Abstract
Translation of the hepatitis C virus (HCV) RNA is mediated by the interaction of ribosomes and cellular proteins with an internal ribosome entry site (IRES) located within the 5'-untranslated region (5'-UTR). We have investigated whether small RNA molecules corresponding to the different stem-loop (SL) domains of the HCV IRES, when introduced in trans, can bind to the cellular proteins and antagonize their binding to the viral IRES, thereby inhibiting HCV IRES-mediated translation. We have found that a RNA molecule corresponding to SL III could efficiently inhibit HCV IRES-mediated translation in a dose-dependent manner without affecting cap-dependent translation. The SL III RNA was found to bind to most of the cellular proteins which interacted with the HCV 5'-UTR. A smaller RNA corresponding to SL e+f of domain III also strongly and selectively inhibited HCV IRES-mediated translation. This RNA molecule interacted with the ribosomal S5 protein and prevented the recruitment of the 40S ribosomal subunit. This study reveals valuable insights into the role of the SL structures of the HCV IRES in mediating ribosome entry. Finally, these results provide a basis for developing anti-HCV therapy using small RNA molecules mimicking the SL structures of the 5'-UTR to specifically block viral RNA translation.
Figures
Figure 1
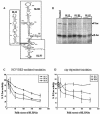
Specific dose-dependent inhibition of HCV IRES-mediated translation in vitro by HCV SL III RNA. (A) Proposed secondary structure of the HCV IRES RNA spanning nucleotides 40–372 of the 5′-UTR of the viral RNA (adapted from 41). The domains that were PCR amplified and cloned to generate small RNAs are delineated. (B) 100- and 200-fold molar excesses of in vitro transcribed SL II, III and IV RNAs were added to in vitro translation reactions of HCV bicistronic RNA. An aliquot of 5 µl of the translation reactions was resolved by SDS–12.5% PAGE and exposed for phosphoimaging. The Fluc and Rluc protein products are indicated by arrows. (C) The percent Fluc activity, representing the efficiency of HCV IRES-mediated translation from a HCV bicistronic template, in the presence of six increasing concentrations of SL II, III and IV RNAs was plotted. The Fluc activity at each concentration is represented as a percentage of the control reaction (expressed as 100%). The data were fitted to a non-linear regression curve to determine IC50 values. (D) The percent Rluc activity, representing the efficiency of cap-dependent translation from the same set of experiments, was plotted. The Rluc activity at each concentration is represented as a percentage of the control reaction. The translation efficiency was not reduced to below 50% by either SL III or SL II.
Figure 2
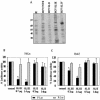
Interaction of HCV 5′-UTR SL RNAs with HeLa cytoplasmic proteins and effect of SL III on HCV IRES-mediated translation in vivo. (A) 32P-labeled RNAs corresponding to the full-length HCV 5′-UTR and SL II, III and IV domains were UV-crosslinked to HeLa S10 cytoplasmic extract, digested with RNase A and resolved by SDS–10% PAGE. M represents 14C-labeled protein molecular weight markers. (B) Three-way co-transfections were performed in HeLa cells using pRL-CMV, pCD-HCV5′-UTR-Fluc and two concentrations of pCD-SL III and pCD-SL II DNAs. DNA quantity per dish was normalized by transfecting pGEM-3Z DNA. The black bars represent Fluc activity (HCV IRES-mediated translation) whereas the gray bars represent Rluc activity (cap-dependent translation). (C) The same experiment was repeated in the Huh7 cell line. Combined data from three independent experiments in each cell line are shown. Luciferase activity in control reactions is expressed as 100%. Values which significantly differ from controls (P < 0.01) are indicated by asterisks.
Figure 3
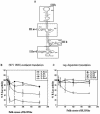
Specific inhibition of HCV IRES-mediated translation in vitro by HCV 5′-UTR SL III e+f RNA. (A) Proposed secondary structure of HCV IRES (internal ribosome entry site) domain III (nucleotides 121–315), delineating the SL structures which were generated by oligonucleotide- driven transcription. (B) The percent Fluc activity, representing the efficiency of HCV IRES-mediated translation from a HCV bicistronic template, in the presence of five increasing concentrations of SL III a+c, b, d and e+f RNAs was plotted. Luciferase activity in control reactions is expressed as 100%. The data for SL III e+f was fitted to a non-linear regression curve to determine the IC50 value. (C) The percent Rluc activity representing the efficiency of cap-dependent translation from the same set of experiments was plotted. The reporter gene activity at each concentration is represented as a percentage of the control reaction.
Figure 4
Effect of SL III e+f RNA on HCV IRES-mediated translation and replication in vivo. (A) Huh7 cells were co-transfected with 6 µg of in vitro transcribed capped HCV bicistronic RNA (schematically represented) and two concentrations (6 and 12 µg) of either SL III e+f RNA or SL III d RNA. The RNA quantities in each dish were normalized by adding appropriate amounts of an in vitro transcribed RNA corresponding to the polylinker sequence of the pGEM 3Z plasmid. The black bars represent Fluc activity (HCV IRES-mediated translation) whereas the gray bars represent Rluc activity (cap-dependent translation). Luciferase activity in control reactions is expressed as 100%. Values which significantly differ from controls (P < 0.01) are indicated by asterisks. (B) Huh7 cells were either mock transfected (lane 1) or transfected with 8 µg of BB7 HCV replicon RNA (schematically represented). Twelve hours post-transfection the cells were retransfected with two concentrations (6 and 12 µg) of SL III e+f RNA (lanes 3 and 4). The cells were harvested 24 h after retransfection, total RNA was isolated and a RNase protection assay was performed using a [32P]UTP-labeled HCV 5′-UTR positive sense probe. The RNAs were resolved by 10% urea–PAGE. Lane M shows the migration of a radiolabeled 100 bp DNA ladder. (C) 100- and 200-fold molar excesses of in vitro transcribed SL III e+f RNA was added to in vitro translation reactions of Fluc-EMCV-GFP bicistronic RNA in RRL. An aliquot of 10 µl of the translation reactions was resolved by SDS–12.5% PAGE and exposed for phosphorimaging. The 35S-labeled Fluc and GFP protein products are indicated by arrows.
Figure 5
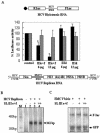
Binding of domain III stem–loop RNAs to HeLa cellular proteins and interaction of SL III e+f RNA with the S5 ribosomal protein. (A) 32P-labeled RNAs corresponding to the HCV SL III a+c, b, d and e+f subdomains were UV-crosslinked to HeLa S10 cytoplasmic extract, digested with RNase A and resolved by SDS–10% PAGE. The position of p25 bound to SL III e+f RNA is indicated by an arrow. (B) Purified recombinant S5 ribosomal protein was UV-crosslinked to HCV SL III e+f, SL III d and full-length HCV 5′-UTR RNA. The nucleoprotein complexes were resolved by SDS–12% PAGE and the position of S5 protein (p25) is indicated.
Figure 6
SL III e+f (A297G) RNA fails to bind to S5 ribosomal protein and does not inhibit HCV IRES-mediated translation. (A) 32P-labeled RNAs corresponding to SL III e+f and SL III e+f (A297G) were UV-crosslinked to HeLa S10 extract and digested with RNase A. The nucleoprotein complexes were resolved by SDS–15% PAGE and the position of p25 is indicated. (B) The same RNAs were UV-crosslinked to purified S5 ribosomal protein and the nucleoprotein complexes were resolved by SDS–15% PAGE. (C) 100- and 200-fold molar excesses of in vitro transcribed SL III e+f (A297G) RNA was added to in vitro translation reactions of HCV bicistronic RNA and luciferase activity was assayed. The black bars represent Fluc activity (HCV IRES-mediated translation) whereas the gray bars represent Rluc activity (cap-dependent translation). Luciferase activity in control reactions is expressed as 100%. Combined data from three independent experiments are represented.
Figure 7
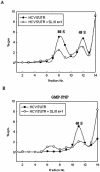
SL III e+f prevents 40s ribosomal subunit recruitment by the HCV IRES. (A) Sucrose gradient sedimentation profiles of [32P]UTP-labeled HCV 5′-UTR RNA incubated in RRL in the absence and presence of a 200-fold excess of unlabeled SL III e+f RNA. (B) Sedimentation profile of radiolabeled HCV 5′-UTR RNA in the presence of 2 mM GMP-PNP in the presence or absence of a 200-fold excess of unlabeled SL III e+f RNA. The filled circles represent the control reaction profile and the open circles show the profile in the presence of SL III e+f. Both profiles show the counts per minute as a percentage of the total counts added to the reaction (∼105 c.p.m.) against the fraction number of the gradient. The fractions were collected from the bottom upwards. The 80S and 48S ribosomal peaks are indicated.
Similar articles
- A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId.
Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S. Kikuchi K, et al. Nucleic Acids Res. 2005 Jan 28;33(2):683-92. doi: 10.1093/nar/gki215. Print 2005. Nucleic Acids Res. 2005. PMID: 15681618 Free PMC article. - Human ribosomal protein L18a interacts with hepatitis C virus internal ribosome entry site.
Dhar D, Mapa K, Pudi R, Srinivasan P, Bodhinathan K, Das S. Dhar D, et al. Arch Virol. 2006 Mar;151(3):509-24. doi: 10.1007/s00705-005-0642-6. Epub 2005 Sep 30. Arch Virol. 2006. PMID: 16195786 - Sequence-specific cleavage of hepatitis C virus RNA by DNAzymes: inhibition of viral RNA translation and replication.
Roy S, Gupta N, Subramanian N, Mondal T, Banerjea AC, Das S. Roy S, et al. J Gen Virol. 2008 Jul;89(Pt 7):1579-1586. doi: 10.1099/vir.0.83650-0. J Gen Virol. 2008. PMID: 18559927 - Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses.
Dasgupta A, Das S, Izumi R, Venkatesan A, Barat B. Dasgupta A, et al. FEMS Microbiol Lett. 2004 May 15;234(2):189-99. doi: 10.1016/j.femsle.2004.03.045. FEMS Microbiol Lett. 2004. PMID: 15135522 Review. - Hepatitis C Virus Translation Regulation.
Niepmann M, Gerresheim GK. Niepmann M, et al. Int J Mol Sci. 2020 Mar 27;21(7):2328. doi: 10.3390/ijms21072328. Int J Mol Sci. 2020. PMID: 32230899 Free PMC article. Review.
Cited by
- Targeting ribosome assembly on the HCV RNA using a small RNA molecule.
Bhat P, Gnanasundram SV, Mani P, Ray PS, Sarkar DP, Das S. Bhat P, et al. RNA Biol. 2012 Aug;9(8):1110-9. doi: 10.4161/rna.21208. Epub 2012 Aug 1. RNA Biol. 2012. PMID: 22858675 Free PMC article. - Inhibition of hepatitis C virus by an M1GS ribozyme derived from the catalytic RNA subunit of Escherichia coli RNase P.
Mao X, Li X, Mao X, Huang Z, Zhang C, Zhang W, Wu J, Li G. Mao X, et al. Virol J. 2014 May 13;11:86. doi: 10.1186/1743-422X-11-86. Virol J. 2014. PMID: 24885776 Free PMC article. - The beta hairpin structure within ribosomal protein S5 mediates interplay between domains II and IV and regulates HCV IRES function.
Bhat P, Shwetha S, Sharma DK, Joseph AP, Srinivasan N, Das S. Bhat P, et al. Nucleic Acids Res. 2015 Mar 11;43(5):2888-901. doi: 10.1093/nar/gkv110. Epub 2015 Feb 24. Nucleic Acids Res. 2015. PMID: 25712089 Free PMC article. - A hepatitis C virus (HCV) internal ribosome entry site (IRES) domain III-IV-targeted aptamer inhibits translation by binding to an apical loop of domain IIId.
Kikuchi K, Umehara T, Fukuda K, Kuno A, Hasegawa T, Nishikawa S. Kikuchi K, et al. Nucleic Acids Res. 2005 Jan 28;33(2):683-92. doi: 10.1093/nar/gki215. Print 2005. Nucleic Acids Res. 2005. PMID: 15681618 Free PMC article.
References
- Jenny-Avital E.R. (1998) Hepatitis C. Curr. Opin. Infect. Dis., 11, 293–299. - PubMed
- Lindsay K.L. (1997) Therapy of hepatitis C: overview. Hepatology, 26, 715–755. - PubMed
- Pudi R., Abhiman,S., Srinivasan,N. and Das,S. (2003) Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by specific interaction of independent regions of human La autoantigen. J. Biol. Chem., 278, 12231–12240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous