A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin - PubMed (original) (raw)
A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin
Gareth J Browne et al. Mol Cell Biol. 2004 Apr.
Abstract
Eukaryotic elongation factor 2 (eEF2) kinase is an unusual calcium- and calmodulin-dependent protein kinase that is regulated by insulin through the rapamycin-sensitive mTOR pathway. Here we show that insulin decreases the ability of eEF2 kinase to bind calmodulin in a rapamycin-sensitive manner. We identify a novel phosphorylation site in eEF2 kinase (Ser78) that is located immediately next to its calmodulin-binding motif. Phosphorylation of this site is increased by insulin in a rapamycin-sensitive fashion. Regulation of the phosphorylation of Ser78 also requires amino acids and the protein kinase phosphoinositide-dependent kinase 1. Mutation of this site to alanine strongly attenuates the effects of insulin and rapamycin both on the binding of calmodulin to eEF2 kinase and on eEF2 kinase activity. Phosphorylation of Ser78 is thus likely to link insulin and mTOR signaling to the control of eEF2 phosphorylation and chain elongation. This site is not a target for known kinases in the mTOR pathway, e.g., the S6 kinases, implying that it is phosphorylated by a novel mTOR-linked protein kinase that serves to couple hormones and amino acids to the control of translation elongation. eEF2 kinase is thus a target for mTOR signaling independently of previously known downstream components of the pathway.
Figures
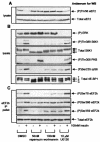
FIG. 1.
The treatment of cells with 2-DOG decreases the phosphorylation of several sites in eEF2 kinase. KB cells (70% confluent) were incubated in DMEM (containing 1 g of
d
-glucose per liter) plus fetal calf serum and treated with 5 or 25 mM 2-DOG for 30 min. (A) Ser78 is located adjacent to the CaM-binding site in eEF2 kinase. The CaM-binding, catalytic, and substrate-binding domains are indicated, as is the amino acid sequence (single-letter code) of the CaM-binding region. Also indicated are other relevant sites of phosphorylation within eEF2 kinase. (B) eEF2 kinase was immunoprecipitated from 100 μg of cell lysate protein and subjected to SDS-PAGE before Western blotting for total eEF2 kinase or phosphorylated eEF2 kinase by using the indicated phosphospecific antisera. (C) A total of 20 μg of cell lysate protein was subjected to SDS-PAGE and Western blotting for phosphorylated and total proteins as indicated. The α to γ species of 4E-BP1 and the different species of S6K1 (which reflect differing states of phosphorylation) are indicated. (D) KB cells (90% confluent) were starved of serum for 16 h and then treated with insulin for 30 min as indicated. eEF2 kinase was immunoprecipitated from 100 μg of cell lysate protein and subjected to SDS-PAGE before Western blotting for total eEF2 kinase or eEF2 kinase phosphorylated at Ser78. (E) KB cells were transfected with vectors encoding wild-type eEF2 kinase or a mutant in which Ser78 had been converted to alanine, each with an epitope (FLAG) tag. Cells were starved of serum and then in some cases treated with insulin (30 min) prior to lysis. The exogenous eEF2 kinase was then immunoprecipitated from the lysates by using anti-FLAG. Immunoprecipitates or cell lysates (as indicated) were then analyzed by SDS-PAGE and Western blotting for total eEF2 kinase, phosphorylated eEF2 kinase using the indicated phosphospecific antisera, or S6K1. eEF2k, eEF2 kinase; WB, Western blotting; WT, wild type; IP, immunoprecipitation.
FIG. 2.
The phosphorylation of Ser78 in eEF2 kinase is regulated by insulin in a rapamycin-sensitive manner. KB cells (90% confluent) were starved of serum for 16 h, pretreated with signaling inhibitors for 30 min, and then treated with insulin for 30 min as indicated. (A) Cell lysate protein (20 μg) was subjected to SDS-PAGE followed by Western blotting for eEF2 phosphorylated at Thr56 or total eEF2. (B) Cell lysate protein (20 μg) was subjected to SDS-PAGE followed by Western blotting for phosphorylated or total proteins as indicated (see also legend to Fig. 1B). (C) Endogenous eEF2 kinase was immunoprecipitated from 100 μg of lysate protein and subjected to SDS-PAGE followed by Western blotting for phosphorylated or total eEF2 kinase, as indicated. eEF2k, eEF2 kinase; WB, Western blotting; DMSO, dimethyl sulfoxide; IP, immunoprecipitation.
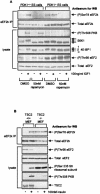
FIG. 3.
The phosphorylation of Ser78 in eEF2 kinase is insensitive to the broad specificity AGC kinase inhibitor Ro31-8220. Serum-starved (16 h) 90% confluent KB cells were pretreated with signaling inhibitors for 30 min and then with insulin for 30 min as indicated. eEF2 kinase was immunoprecipitated from 100 μg of lysate (IP pellet), or 20 μg of cell lysate protein was analyzed directly. Samples were subjected to SDS-PAGE followed by Western blotting (WB) for phosphorylated or total proteins, as indicated. eEF2k, eEF2 kinase.
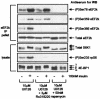
FIG. 4.
Phosphorylation of Ser78 in eEF2 kinase is sensitive to amino acid withdrawal. (A) CHO.K1 cells (90% confluent) were starved of serum for 16 h, preincubated for 1 h in serum-free Ham's F12 medium, Dulbecco's phosphate-buffered saline (D-PBS) containing 1 g of glucose per liter, or the latter containing 1 g of
d
-glucose per liter plus amino acids (AA) prior to treatment with insulin for 30 min as indicated. Where used, CHX was added 30 min prior to insulin stimulation. eEF2 kinase was immunoprecipitated from 100 μg of lysate protein or 20 μg of cell lysate protein and subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. (B) The method was the same as in panel A except that KB cells were used and were starved of amino acids for 4 h in Earle's balanced salts solution (EBSS); no treatments with CHX were included. Samples were analyzed as in panel A, except that Western blotting was also performed for phosphorylated rpS6 (Ser235) and for phosphorylation of Ser366 and Ser359 in eEF2 kinase. WB, Western blotting; IP, immunoprecipitation; eEF2k, eEF2 kinase.
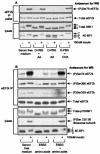
FIG. 5.
Phosphorylation of Ser78 in eEF2 kinase is dysregulated in cells lacking PDK1 or TSC2. (A) Ninety percent confluent ES cells (PDK1+/+ or PDK1−/− as indicated) were serum starved for 3 h and then pretreated with rapamycin (or dimethyl sulfide [DMSO] as a control) for 30 min prior to IGF1 treatment for 30 min. eEF2 kinase immunoprecipitated from 500 μg of cell lysate protein or 20 μg of total cell lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. The positions of the differently phosphorylated forms of S6K1 and 4E-BP1 are indicated by arrows. (B) Ninety percent confluent mouse embryo fibroblasts (TSC2+/+ or TSC2−/− as indicated) were serum starved for 3 h and then, where indicated, treated with 100 nM insulin for 30 min. eEF2 kinase that had been immunoprecipitated from 500 μg of cell lysate protein or 20 μg of total cell lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated and total proteins as indicated. eEF2k, eEF2 kinase; WB, Western blotting; IP, immunoprecipitation.
FIG. 6.
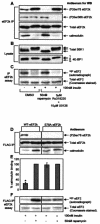
Ser78 is required for the regulation of the binding of CaM to eEF2 kinase by insulin. (A to C) KB cells (90% confluent) were serum starved for 16 h and then pretreated with the indicated signaling inhibitors for 30 min prior to treatment with insulin for 30 min as indicated. The cells were lysed, and immunoprecipitations and eEF2 kinase assays were carried out in a modified extraction buffer containing 1 mM CaCl2 as described in Materials and Methods. In panel A, eEF2 kinase immunoprecipitated from 100 μg of lysate protein was subjected to SDS-PAGE followed by Western blotting for phosphorylated or total eEF2 kinase as indicated. In panel B, cell lysate protein (20 μg) was analyzed by SDS-PAGE and Western blotting using antisera for S6K1 or 4E-BP1, as indicated. Positions of the differently phosphorylated forms of these proteins are shown. In panel C, eEF2 kinase immunoprecipitated from 100 μg of cell lysate protein was incubated with 1 μg of purified eEF2 in the presence of [γ-32P]ATP. Samples were then subjected to SDS-PAGE. Gels were stained with Coomassie and analyzed by autoradiography to assess incorporation of radiolabel into eEF2 (i.e., eEF2 kinase activity, upper section). The lower section shows a portion of the stained gel to confirm equal loading of the substrate, eEF2. (D to F) KB cells were transfected with vectors encoding FLAG-tagged versions of wild-type eEF2 kinase or the Ser78Ala mutant. When cells had reached 90% confluence, they were starved of serum for 16 h and then pretreated with rapamycin for 30 min prior to treatment with insulin as indicated. The FLAG-tagged eEF2 kinase was immunoprecipitated from 100 μg of lysate protein by using immobilized FLAG antibody and subjected to SDS-PAGE followed by Western blotting for phosphorylated and total eEF2 kinase or bound CaM as indicated in panel D. The graph in panel E shows the binding of CaM to wild-type FLAG-eEF2 kinase or the Ser78Ala mutant immunoprecipitated from cells treated with insulin or insulin plus rapamycin. The amount of bound CaM, presented as a percentage of the CaM bound to the FLAG-eEF2 kinase immunoprecipitated from cells treated with insulin plus rapamycin, was determined by using Image/J software (available at rsb.info.nih.gov/ij/). The results are presented as the means ± standard errors of the means (n = 5). Alternatively, in panel F eEF2 kinase activity was determined by incubating the immunoprecipitated eEF2 kinase with 1 μg of purified eEF2 in the presence of [γ-32P]ATP to assay eEF2 kinase activity (see Materials and Methods) (40). Samples were then subjected to SDS-PAGE. Gels were stained with Coomassie brilliant blue and then analyzed by autoradiography to assess incorporation of the label into eEF2 (upper section). The lower section shows a portion of the stained gel to confirm equal loading of eEF2, the substrate. eEF2k, eEF2 kinase; WB, Western blotting; IP, immunoprecipitation; WT, wild type; *, P < 0.005.
Similar articles
- The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses.
Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. Smith EM, et al. J Biol Chem. 2005 May 13;280(19):18717-27. doi: 10.1074/jbc.M414499200. Epub 2005 Mar 16. J Biol Chem. 2005. PMID: 15772076 - mTOR-mediated regulation of translation factors by amino acids.
Proud CG. Proud CG. Biochem Biophys Res Commun. 2004 Jan 9;313(2):429-36. doi: 10.1016/j.bbrc.2003.07.015. Biochem Biophys Res Commun. 2004. PMID: 14684180 Review. - Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase.
Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Wang X, et al. EMBO J. 2001 Aug 15;20(16):4370-9. doi: 10.1093/emboj/20.16.4370. EMBO J. 2001. PMID: 11500364 Free PMC article. - Myocardial ischemia and increased heart work modulate the phosphorylation state of eukaryotic elongation factor-2.
Horman S, Beauloye C, Vertommen D, Vanoverschelde JL, Hue L, Rider MH. Horman S, et al. J Biol Chem. 2003 Oct 24;278(43):41970-6. doi: 10.1074/jbc.M302403200. Epub 2003 Aug 14. J Biol Chem. 2003. PMID: 12920134 - Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation.
Kaul G, Pattan G, Rafeequi T. Kaul G, et al. Cell Biochem Funct. 2011 Apr;29(3):227-34. doi: 10.1002/cbf.1740. Epub 2011 Mar 10. Cell Biochem Funct. 2011. PMID: 21394738 Review.
Cited by
- mTORC1 and mTORC2 in cancer and the tumor microenvironment.
Kim LC, Cook RS, Chen J. Kim LC, et al. Oncogene. 2017 Apr 20;36(16):2191-2201. doi: 10.1038/onc.2016.363. Epub 2016 Oct 17. Oncogene. 2017. PMID: 27748764 Free PMC article. Review. - Alcohol and NMDA receptor: current research and future direction.
Chandrasekar R. Chandrasekar R. Front Mol Neurosci. 2013 May 28;6:14. doi: 10.3389/fnmol.2013.00014. eCollection 2013. Front Mol Neurosci. 2013. PMID: 23754976 Free PMC article. - Regulation of mRNA translation by signaling pathways.
Roux PP, Topisirovic I. Roux PP, et al. Cold Spring Harb Perspect Biol. 2012 Nov 1;4(11):a012252. doi: 10.1101/cshperspect.a012252. Cold Spring Harb Perspect Biol. 2012. PMID: 22888049 Free PMC article. Review. - Nutritionally essential amino acids and metabolic signaling in aging.
Dillon EL. Dillon EL. Amino Acids. 2013 Sep;45(3):431-41. doi: 10.1007/s00726-012-1438-0. Epub 2012 Dec 14. Amino Acids. 2013. PMID: 23239011 Free PMC article. Review. - IGF-I activates the eIF4F system in cardiac muscle in vivo.
Vary TC, Lang CH. Vary TC, et al. Mol Cell Biochem. 2005 Apr;272(1-2):209-20. doi: 10.1007/s11010-005-7551-6. Mol Cell Biochem. 2005. PMID: 16010989
References
- Alessi, D. R. 1997. The protein kinase C inhibitors Ro31-8220 and GF109203X are equally potent inhibitors of MAPKAP kinase-1β and p70 S6 kinase. FEBS Lett. 402:121-123. - PubMed
- Alessi, D. R., M. T. Kozlowski, Q.-P. Weng, N. Morrice, and J. Avruch. 1998. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol. 8:69-81. - PubMed
- Avruch, J., C. Belham, Q. Weng, K. Hara, and K. Yonezawa. 2001. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 26:115-154. - PubMed
- Balendran, A., G. R. Hare, A. Kieloch, M. R. Williams, and D. R. Alessi. 2000. Further evidence that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is required for the stability and phosphorylation of protein kinase C (PKC) isoforms. FEBS Lett. 484:217-223. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous