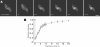Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis - PubMed (original) (raw)
Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis
Yuda Fang et al. Mol Biol Cell. 2004 Jun.
Abstract
The organization of the pre-mRNA splicing machinery has been extensively studied in mammalian and yeast cells and far less is known in living plant cells and different cell types of an intact organism. Here, we report on the expression, organization, and dynamics of pre-mRNA splicing factors (SR33, SR1/atSRp34, and atSRp30) under control of their endogenous promoters in Arabidopsis. Distinct tissue-specific expression patterns were observed, and differences in the distribution of these proteins within nuclei of different cell types were identified. These factors localized in a cell type-dependent speckled pattern as well as being diffusely distributed throughout the nucleoplasm. Electron microscopic analysis has revealed that these speckles correspond to interchromatin granule clusters. Time-lapse microscopy revealed that speckles move within a constrained nuclear space, and their organization is altered during the cell cycle. Fluorescence recovery after photobleaching analysis revealed a rapid exchange rate of splicing factors in nuclear speckles. The dynamic organization of plant speckles is closely related to the transcriptional activity of the cells. The organization and dynamic behavior of speckles in Arabidopsis cell nuclei provides significant insight into understanding the functional compartmentalization of the nucleus and its relationship to chromatin organization within various cell types of a single organism.
Figures
Figure 1.
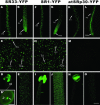
Expression and nuclear localization patterns of SR33-YFP, SR1/atSRp34-YFP, and atSRp30-YFP in Arabidopsis. Projections of a series of confocal optical sections of inflorescence stems, anthers, roots, and leaves are shown. (A, F, and K) In the basal section of primary root. (B, G, and L) In the tip section of primary root. (C, H, and M) In leaf. (D, I, and N) In anther (D′, enlarged view of a pollen grain, arrow highlights the vegetative nucleus and arrowheads highlight the nuclei of sperm cells; bar, 10 μm). (E, J, and O) In inflorescence stem. Different cell types are highlighted with arrows: Gu, guard cells; LEP, larger (C) and small (H) leaf epidermal pavement cells; RE, root epidermal cells; RH, root hairs; RM, root meristematic cells; Tri, trichomes. Bar, 100 μm.
Figure 2.
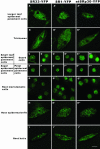
Localization of SR33-YFP, SR1/atSRp34-YFP, and atSRp30-YFP in different Arabidopsis cell types. Maximum projections of deconvolved optical sections are shown. The localization patterns of the three SR-YFP fusions are different among cell types with variable nuclear sizes and shapes. Bar, 5 μm.
Figure 3.
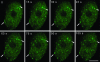
Immunoelectron microscopic localization of SR proteins in Arabidopsis. Root sections are immunolabeled with 3C5 antibody, which recognizes the SR family of pre-mRNA splicing factors in IGCs (arrows). Chr, chromatin; No, nucleolus. Bar, 200 nm.
Figure 4.
Time-lapse deconvolution microscopy of SR1/atSRp34-YFP in a living leaf epidermal pavement cell. The projections of deconvolved optical sections are shown at each time point. The arrows highlight the fusion and disassociation of two speckles, and the arrowheads highlight the splicing factors being released from speckles. The time is indicated in seconds. Bar, 5 μm.
Figure 5.
FRAP of SR proteins. (A) Leaf epidermal pavement cells expressing SR1/atSRp34-YFP were imaged before and during recovery from photobleaching. The white square frames represent the photobleached region. Bar, 5 μm. (B) Kinetics of recovery after bleaching of SR1/atSRp34-YFP. * indicates the photobleach point.
Figure 6.
Dynamics of SR1/atSRp34-YFP during mitosis in living root epidermal cells. (A) A cell (arrow) just before nuclear envelope breakdown. (B and C) In prophase, as the nuclear envelope breaks down, the splicing factors enter the cytoplasm. (D) In metaphase, splicing factors are diffusely distributed in the cytoplasm. (E–J) Splicing factors are reentering into daughter nuclei. Newly forming speckles are observed in telophase nuclei (arrows in F–I). Bar, 5 μm.
Figure 7.
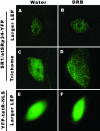
Effect of transcription inhibition on the organization of SR proteins. The transgenic leaves were treated with 50 μg/ml DRB for 5 h at 22°C. In leaf epidermal pavement cells (B), after transcriptional inhibition, the splicing factors further accumulated in speckles with less labeling in the nucleoplasm. In trichomes (D), after transcriptional inhibition, the splicing factors organized into thousands of microspeckles. DRB (F) has no obvious effect on nuclear morphology and the diffuse distribution of a control protein YFP-tetR-NLS. In control samples, water treatment for 5 hat 22°C had no obvious effect on the distribution of SR-YFP or YFP-tetR-NLS (A, C, and E). Bar, 5 μm.
Similar articles
- Insights into nuclear organization in plants as revealed by the dynamic distribution of Arabidopsis SR splicing factors.
Tillemans V, Leponce I, Rausin G, Dispa L, Motte P. Tillemans V, et al. Plant Cell. 2006 Nov;18(11):3218-34. doi: 10.1105/tpc.106.044529. Epub 2006 Nov 17. Plant Cell. 2006. PMID: 17114353 Free PMC article. - Functional distribution and dynamics of Arabidopsis SR splicing factors in living plant cells.
Tillemans V, Dispa L, Remacle C, Collinge M, Motte P. Tillemans V, et al. Plant J. 2005 Feb;41(4):567-82. doi: 10.1111/j.1365-313X.2004.02321.x. Plant J. 2005. PMID: 15686520 - Co-localisation studies of Arabidopsis SR splicing factors reveal different types of speckles in plant cell nuclei.
Lorković ZJ, Hilscher J, Barta A. Lorković ZJ, et al. Exp Cell Res. 2008 Oct 15;314(17):3175-86. doi: 10.1016/j.yexcr.2008.06.020. Epub 2008 Jul 2. Exp Cell Res. 2008. PMID: 18674533 - Nuclear speckles.
Spector DL, Lamond AI. Spector DL, et al. Cold Spring Harb Perspect Biol. 2011 Feb 1;3(2):a000646. doi: 10.1101/cshperspect.a000646. Cold Spring Harb Perspect Biol. 2011. PMID: 20926517 Free PMC article. Review. - Nuclear speckles: a model for nuclear organelles.
Lamond AI, Spector DL. Lamond AI, et al. Nat Rev Mol Cell Biol. 2003 Aug;4(8):605-12. doi: 10.1038/nrm1172. Nat Rev Mol Cell Biol. 2003. PMID: 12923522 Review.
Cited by
- LACHESIS restricts gametic cell fate in the female gametophyte of Arabidopsis.
Gross-Hardt R, Kägi C, Baumann N, Moore JM, Baskar R, Gagliano WB, Jürgens G, Grossniklaus U. Gross-Hardt R, et al. PLoS Biol. 2007 Mar;5(3):e47. doi: 10.1371/journal.pbio.0050047. PLoS Biol. 2007. PMID: 17326723 Free PMC article. - Nucleolus-tethering system (NoTS) reveals that assembly of photobodies follows a self-organization model.
Liu Y, Liu Q, Yan Q, Shi L, Fang Y. Liu Y, et al. Mol Biol Cell. 2014 Apr;25(8):1366-73. doi: 10.1091/mbc.E13-09-0527. Epub 2014 Feb 19. Mol Biol Cell. 2014. PMID: 24554768 Free PMC article. - Nuclear activity of sperm cells during Hyacinthus orientalis L. in vitro pollen tube growth.
Zienkiewicz K, Suwinska A, Niedojadło K, Zienkiewicz A, Bednarska E. Zienkiewicz K, et al. J Exp Bot. 2011 Jan;62(3):1255-69. doi: 10.1093/jxb/erq354. Epub 2010 Nov 16. J Exp Bot. 2011. PMID: 21081664 Free PMC article. - Regulation of phase separation and antiviral activity of Cactin by glycolytic enzyme PGK via phosphorylation in Drosophila.
Chen D, Shi C, Xu W, Rong Q, Wu Q. Chen D, et al. mBio. 2024 Apr 10;15(4):e0137823. doi: 10.1128/mbio.01378-23. Epub 2024 Mar 6. mBio. 2024. PMID: 38446061 Free PMC article. - Maize rough endosperm3 encodes an RNA splicing factor required for endosperm cell differentiation and has a nonautonomous effect on embryo development.
Fouquet R, Martin F, Fajardo DS, Gault CM, Gómez E, Tseung CW, Policht T, Hueros G, Settles AM. Fouquet R, et al. Plant Cell. 2011 Dec;23(12):4280-97. doi: 10.1105/tpc.111.092163. Epub 2011 Dec 2. Plant Cell. 2011. PMID: 22138152 Free PMC article.
References
- Acevedo, R., Samaniego, R., and Moreno Díaz de la Espina, S. (2002). Coiled bodies in nuclei from plant cells evolving from dormancy to proliferation. Chromosoma 110, 559-569. - PubMed
- Ali, G.S., Golovkin, M., and Reddy, A.S. (2003). Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J. 36, 883-893. - PubMed
- Baluska, F. (1990). Nuclear size, DNA content, and chromatin condensation are different in individual tissues of the maize apex. Protoplasma 158, 1-2.
- Beven, A.F., Simpson, G.G., Brown, J.W.S., and Shaw, P.J. (1995). The organization of splicesomal components in the nuclei of higher plants. J. Cell Sci. 108, 509-518. - PubMed
- Boudonck, K., Dolan, L., and Shaw, P.J. (1998). Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cycle parameters. J. Cell Sci. 111, 3687-3694. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials