SARS--beginning to understand a new virus - PubMed (original) (raw)
Review
SARS--beginning to understand a new virus
Konrad Stadler et al. Nat Rev Microbiol. 2003 Dec.
Abstract
The 114-day epidemic of the severe acute respiratory syndrome (SARS) swept 29 countries, affected a reported 8,098 people, left 774 patients dead and almost paralyzed the Asian economy. Aggressive quarantine measures, possibly aided by rising summer temperatures, successfully terminated the first eruption of SARS and provided at least a temporal break, which allows us to consolidate what we have learned so far and plan for the future. Here, we review the genomics of the SARS coronavirus (SARS-CoV), its phylogeny, antigenic structure, immune response and potential therapeutic interventions should the SARS epidemic flare up again.
Conflict of interest statement
S. Abrignani, R. Rappuoli, K. Stadler and V. Masignani are employed by Chiron Corporation.
Figures
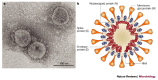
Figure 1. Morphology of the SARS coronavirus.
a | Electron micrograph of the virus that was cultivated in Vero cells (Image courtesy of Dr L. Kolesnikova, Institute of Virology, Marburg, Germany). Large, club-shaped protrusions consisting of spike protein form a crown-like corona that gives the virus its name. b | Schematic representation of the virus. A lipid bilayer comprising the spike protein, the membrane glycoprotein and the envelope protein cloaks the helical nucleocapsid, which consists of the nucleocapsid protein that is associated with the viral RNA. In the case of coronaviruses, the lipid envelope is derived from intracellular membranes.
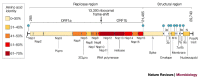
Figure 2. Genome structure of SARS coronavirus.
Replicase and structural regions are shown together with the predicted cleavage products in ORF1a and ORF1b. The position of the leader sequence (L), the 3′ poly(A) tract and the ribosomal frameshift site between ORF1a and ORF1b are also indicated. Each box represents a protein product (Nsp, non-structural protein). Colours indicate the level of amino-acid identity with the best-matching protein of other coronaviruses (Table 2). The SARS-CoV accessory genes are white. Filled circles indicate the positions of the nine transcription-regulatory sequences (TRSs) that are specific for SARS-CoV (5′ACGAAC3′).
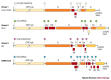
Figure 3. Comparison of coronavirus genome structures.
Genome organization of coronavirus representatives of group 1 (human coronavirus 229E, HCoV-229E), group 2 (mouse hepatitis virus, MHV) and group 3 (avian infectious bronchitis virus, IBV; SARS-CoV). Red boxes represent the accessory genes. The positions of the leader sequence (L) and poly(A) tract are indicated; circles of different colour represent group-specific transcription-regulatory sequences (TRS).
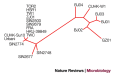
Figure 4. Molecular relationship of 20 SARS genomes.
The unrooted tree was obtained through the alignment of whole-genome sequences considering only sequence variants that occurred at least twice. The analysis was performed using the maximum likelihood criterion as implemented in the Phylip package.
Figure 5. Relationship between SARS-CoV and other coronaviruses using different phylogenetic strategies.
a | Unrooted tree obtained by comparing the well-conserved polymerase protein sequence. According to this approach, SARS-CoV belongs to a new group. The tree has been constructed using the protein sequences of the RNA-dependent RNA polymerase of the following coronaviruses: porcine epidemic diarrhea virus (PEDV), human coronavirus 229E (HCoV-229E), canine coronavirus (CCV), feline infectious peritonitis virus (FIPV), transmissible gastroenteritis virus (TGV), mouse hepatitis virus (MHV), bovine coronavirus (BCoV), syaloacryoadenitis virus of rats (SDAV), human coronavirus OC43 (OC43), haemagglutinating encephalomyelitis virus of swine (PHEV), turkey coronavirus (TCV), avian infectious bronchitis virus (IBV) and SARS-CoV. b | Tree obtained using the sequence of the S1 domain of the spike protein. The multiple sequence alignment was constructed using consensus sequences generated from group 1 and group 2 coronaviruses (G1 cons and G2 cons), the sequence of IBV (group 3) and of SARS-CoV. The neighbour-joining algorithm was used to build the tree. Numbers represent the result of a bootstrap analysis performed with 100 replicates.
Figure 6. The S1 domain of SARS-CoV spike is structurally related to group 2 coronaviruses.
Schematic representation of cysteine positions in the S1 domains of group 1, 2 and 3 coronaviruses, compared with the SARS-CoV spike protein. Horizontal bars represent the S1 amino-acid sequences (in the case of SARS-CoV and IBV) or the consensus profiles (generated from group 1, (G1 cons) and from group 2 (G2 cons)). The bars are drawn to scale. Relative cysteine positions are indicated by rectangular bars. Only cysteines that are conserved within each consensus are reported. Coloured lines connect cysteines that are conserved between the SARS-CoV S1 domain and the consensus sequence generated from the group 1 (green), group 2 (red) and IBV S1 sequences (blue).
Similar articles
- SARS: understanding the virus and development of rational therapy.
Stadler K, Rappuoli R. Stadler K, et al. Curr Mol Med. 2005 Nov;5(7):677-97. doi: 10.2174/156652405774641124. Curr Mol Med. 2005. PMID: 16305493 Review. - Molecular biology of severe acute respiratory syndrome coronavirus.
Ziebuhr J. Ziebuhr J. Curr Opin Microbiol. 2004 Aug;7(4):412-9. doi: 10.1016/j.mib.2004.06.007. Curr Opin Microbiol. 2004. PMID: 15358261 Free PMC article. Review. - Virology. The SARS coronavirus: a postgenomic era.
Holmes KV, Enjuanes L. Holmes KV, et al. Science. 2003 May 30;300(5624):1377-8. doi: 10.1126/science.1086418. Science. 2003. PMID: 12775826 No abstract available. - Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus.
Tan YJ, Lim SG, Hong W. Tan YJ, et al. Antiviral Res. 2006 Nov;72(2):78-88. doi: 10.1016/j.antiviral.2006.05.010. Epub 2006 Jun 6. Antiviral Res. 2006. PMID: 16820226 Free PMC article. Review. - SARS virus: the beginning of the unraveling of a new coronavirus.
Lai MM. Lai MM. J Biomed Sci. 2003 Nov-Dec;10(6 Pt 2):664-75. doi: 10.1159/000074077. J Biomed Sci. 2003. PMID: 14631105 Free PMC article. Review.
Cited by
- Solution structure of mouse hepatitis virus (MHV) nsp3a and determinants of the interaction with MHV nucleocapsid (N) protein.
Keane SC, Giedroc DP. Keane SC, et al. J Virol. 2013 Mar;87(6):3502-15. doi: 10.1128/JVI.03112-12. Epub 2013 Jan 9. J Virol. 2013. PMID: 23302895 Free PMC article. - Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses.
Chen Y, Zhao X, Zhou H, Zhu H, Jiang S, Wang P. Chen Y, et al. Nat Rev Immunol. 2023 Mar;23(3):189-199. doi: 10.1038/s41577-022-00784-3. Epub 2022 Sep 27. Nat Rev Immunol. 2023. PMID: 36168054 Free PMC article. Review. - A deep ensemble learning-based automated detection of COVID-19 using lung CT images and Vision Transformer and ConvNeXt.
Tian G, Wang Z, Wang C, Chen J, Liu G, Xu H, Lu Y, Han Z, Zhao Y, Li Z, Luo X, Peng L. Tian G, et al. Front Microbiol. 2022 Nov 4;13:1024104. doi: 10.3389/fmicb.2022.1024104. eCollection 2022. Front Microbiol. 2022. PMID: 36406463 Free PMC article. - Antiviral applications of RNAi for coronavirus.
Wu CJ, Chan YL. Wu CJ, et al. Expert Opin Investig Drugs. 2006 Feb;15(2):89-97. doi: 10.1517/13543784.15.2.89. Expert Opin Investig Drugs. 2006. PMID: 16433589 Free PMC article. Review. - Cellular entry of the SARS coronavirus.
Hofmann H, Pöhlmann S. Hofmann H, et al. Trends Microbiol. 2004 Oct;12(10):466-72. doi: 10.1016/j.tim.2004.08.008. Trends Microbiol. 2004. PMID: 15381196 Free PMC article. Review.
References
- WHO. Acute respiratory syndrome in China. [online], (cited 15 Oct 2003), <http://www.who.int/csr/don/2003_2_20/en/> (2003).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous