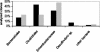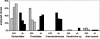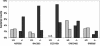An abundance of Escherichia coli is harbored by the mucosa-associated bacterial flora of interleukin-2-deficient mice - PubMed (original) (raw)
An abundance of Escherichia coli is harbored by the mucosa-associated bacterial flora of interleukin-2-deficient mice
M Schuppler et al. Infect Immun. 2004 Apr.
Abstract
Mice deficient in interleukin-2 are well suited for use as an animal model for inflammatory bowel disease. Raised under specific-pathogen-free conditions, interleukin-2-deficient mice develop an inflammatory bowel disease resembling ulcerative colitis in humans. The finding that colitis was attenuated when the mice were kept under germfree conditions implies that the resident intestinal flora is involved in the pathogenesis of colitis. The present study addresses the composition of the mucosa-associated bacterial flora in colon samples from interleukin-2-deficient mice that developed colitis. This was investigated by comparative 16S ribosomal DNA (rDNA) sequence analysis and fluorescence in situ hybridization using rRNA-targeted fluorescent probes to quantify the bacterial populations of the mucosa-associated flora. The investigations revealed distinct differences in the bacterial composition of the mucosa-associated flora between interleukin-2-deficient mice and healthy controls. Fluorescence in situ hybridization identified up to 10% of the mucosa-associated flora in interleukin-2-deficient mice as Escherichia coli, whereas no E. coli was detected in the mucosa from healthy wild-type mice. This finding was consistent with the results from comparative 16S rDNA analysis. About one-third of the clones analyzed from 16S rDNA libraries of interleukin-2-deficient mice represented Enterobacteriaceae, whereas none of the clones analyzed from the healthy controls harbored 16S rDNA from Enterobacteriaceae. The abundance of E. coli in the colonic mucosa of interleukin-2-deficient mice strongly suggests a participation in the pathogenesis of colitis in the interleukin-2-deficient mouse model for inflammatory bowel disease.
Figures
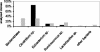
FIG. 1.
Effects of different primers on the composition of the resulting 16S rDNA clone libraries. The broad-range primer pairs 27F-1492R (solid bars) and 63F-1387R (shaded bars) were used to generate two different 16S rDNA clone libraries from the same colon sample of a wild-type control mouse. The resulting 16S rDNA clone libraries were analyzed by comparative sequence analysis of about 60 randomly chosen recombinant clones. Each bar indicates the percentage of clones representing a specific bacterial group in relation to the total number of clones analyzed.
FIG. 2.
Bacterial composition in 16S rDNA clone libraries from proximal (solid bars) and distal (shaded bars) parts of the colon from an IL-2-deficient mouse. Bars indicate the percentages of sequences from Bacteroidales, Clostridiales, Enterobacteriaceae, Desulfovibrio, and other bacteria in relation to total clones analyzed. The analysis was performed for 60 recombinant clones from each of the two clone libraries.
FIG. 3.
Bacterial composition of the mucosa-associated flora determined by comparative 16S rDNA analysis of wild-type control mice and IL-2-deficient mice. 16S rDNA clone libraries were established from three wild-type control mice (+/+) (shaded bars) and three IL-2-deficient mice (−/−) (solid bars). About 60 randomly chosen clones of each library were sequenced and compared to databases. The amount of each bacterial subgroup is expressed as a percentage of the total number of analyzed clones represented in the 16S rDNA clone library for each mouse.
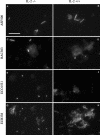
FIG.4.
Whole-cell in situ hybridization of mucosa-associated microflora from IL-2-deficient mice (−/−) (left) and wild-type control mice (+/+) (right) by use of the fluorescent rRNA-targeted oligonucleotide probes shown. Bar, 20 μm (applies to all photomicrographs). (A and B) Probe ASF500 identified tapered rods as part of the mucosa-associated microflora in both groups of mice. (C and D) Probe BAC303 facilitated the specific detection of members of Bacteroides and Prevotella in all samples analyzed. (E and F) Probe ECO1531 identified E. coli cells in samples from IL-2-deficient mice, whereas no signals were observed in samples from wild-type mice. (G and H) The _Bacteria_-specific probe EUB338 was used as a positive control to prove that bacteria were accessible for the fluorescent rRNA-targeted oligonucleotide probes used.
FIG. 5.
Quantification of bacterial populations in colon samples from two healthy wild-type control mice (+/+) (shaded bars) and two IL-2-deficient mice (−/−) (solid bars) by whole-cell hybridization using a set of fluorescent rRNA-targeted oligonucleotide probes. The specificities of the oligonucleotide probes used are shown in Table 1. Each bar represents an individual animal and reflects the mean percentage of specific bacterial populations relative to the total bacterial counts from two independent hybridizations. n.d., no bacteria detected.
Similar articles
- Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum.
Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Baumgart M, et al. ISME J. 2007 Sep;1(5):403-18. doi: 10.1038/ismej.2007.52. Epub 2007 Jul 12. ISME J. 2007. PMID: 18043660 - Dynamics of the mucosa-associated flora in ulcerative colitis patients during remission and clinical relapse.
Ott SJ, Plamondon S, Hart A, Begun A, Rehman A, Kamm MA, Schreiber S. Ott SJ, et al. J Clin Microbiol. 2008 Oct;46(10):3510-3. doi: 10.1128/JCM.01512-08. Epub 2008 Aug 13. J Clin Microbiol. 2008. PMID: 18701655 Free PMC article. - Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease.
Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Mylonaki M, et al. Inflamm Bowel Dis. 2005 May;11(5):481-7. doi: 10.1097/01.mib.0000159663.62651.4f. Inflamm Bowel Dis. 2005. PMID: 15867588 - The bacterial flora in inflammatory bowel disease: current insights in pathogenesis and the influence of antibiotics and probiotics.
Linskens RK, Huijsdens XW, Savelkoul PH, Vandenbroucke-Grauls CM, Meuwissen SG. Linskens RK, et al. Scand J Gastroenterol Suppl. 2001;(234):29-40. doi: 10.1080/003655201753265082. Scand J Gastroenterol Suppl. 2001. PMID: 11768558 Review. - Colonic bacterial flora: changing understandings in the molecular age.
Mai V, Morris JG Jr. Mai V, et al. J Nutr. 2004 Feb;134(2):459-64. doi: 10.1093/jn/134.2.459. J Nutr. 2004. PMID: 14747689 Review.
Cited by
- Inhibitory effects of fermented brown rice on induction of acute colitis by dextran sulfate sodium in rats.
Kataoka K, Ogasa S, Kuwahara T, Bando Y, Hagiwara M, Arimochi H, Nakanishi S, Iwasaki T, Ohnishi Y. Kataoka K, et al. Dig Dis Sci. 2008 Jun;53(6):1601-8. doi: 10.1007/s10620-007-0063-3. Dig Dis Sci. 2008. PMID: 17957470 - Microbial abundance on the eggs of a passerine bird and related fitness consequences between urban and rural habitats.
Lee SI, Lee H, Jablonski PG, Choe JC, Husby M. Lee SI, et al. PLoS One. 2017 Sep 27;12(9):e0185411. doi: 10.1371/journal.pone.0185411. eCollection 2017. PLoS One. 2017. PMID: 28953940 Free PMC article. - Dextran sodium sulfate-induced inflammation alters the expression of proteins by intestinal Escherichia coli strains in a gnotobiotic mouse model.
Schumann S, Alpert C, Engst W, Loh G, Blaut M. Schumann S, et al. Appl Environ Microbiol. 2012 Mar;78(5):1513-22. doi: 10.1128/AEM.07340-11. Epub 2011 Dec 30. Appl Environ Microbiol. 2012. PMID: 22210207 Free PMC article. - Mechanisms of intestinal inflammation and development of associated cancers: lessons learned from mouse models.
Westbrook AM, Szakmary A, Schiestl RH. Westbrook AM, et al. Mutat Res. 2010 Jul-Sep;705(1):40-59. doi: 10.1016/j.mrrev.2010.03.001. Epub 2010 Mar 16. Mutat Res. 2010. PMID: 20298806 Free PMC article. Review.
References
- Amann, R., W. Ludwig, and K. H. Schleifer. 1992. Identification and in situ detection of individual bacterial cells. FEMS Microbiol. Lett. 100:45-50.
- Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases