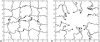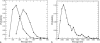Metastable network model of protein transport through nuclear pores - PubMed (original) (raw)
Metastable network model of protein transport through nuclear pores
T Kustanovich et al. Biophys J. 2004 Apr.
Abstract
To reconcile the observed selectivity and the high rate of translocation of cargo-importin complexes through nuclear pores, we propose that the core of the nuclear pore complex is blocked by a metastable network of phenylalanine and glycine nucleoporins. Although the network arrests the unfacilitated passage of objects larger than its mesh size, cargo-importin complexes act as catalysts that reduce the free energy barrier between the cross-linked and the dissociated states of the Nups, and open the network. Using Brownian dynamics simulations we calculate the distribution of passage times through the network for inert particles and cargo-importin complexes of different sizes and discuss the implications of our results for experiments on translocation of proteins through the nuclear pore complex.
Figures
FIGURE 1
A snapshot of network fluctuations (a) and of the grafted chains (b) at T = 0.3. The beads shown are those with _c_-c (chain-chain) interaction.
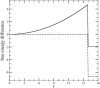
FIGURE 2
The free energy of a chain versus the distance r of its free end from the wall. The combined contribution from the binding energy due to _c_-c interactions (dashed line) and from the elastic entropy (a parabolic curve) is given by solid line. Notice that the barrier height is much higher than the equilibrium binding free energy.
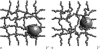
FIGURE 3
An IT (a) and CIC (b) with _R_t = 4 can pass through the network, the former due to thermal fluctuations (in a fully connected network ξ = 7.2) and the latter by also locally breaking the mesh.
FIGURE 4
CIC with _R_t = 8 passes through the network by breaking it.
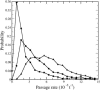
FIGURE 5
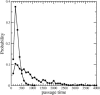
Histogram of passage rates of IT with _R_t = 3 (▴), _R_t = 3.5 (•), and _R_t = 4 (▪). The passage rate is given in units of (1000 × t)−1.
FIGURE 6
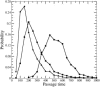
Histogram of passage times of IT (•) and CIC with _N_t = 6 (▪), both of size _R_t = 4.
FIGURE 7
Histogram of passage times for IT with _R_t = 3 (▴), and for CICs with, _R_t = 4, _N_t = 6 (▪); and _R_t = 8, _N_t = 6 (•).
FIGURE 8
Histogram of passage times for IT with (a) _R_t = 4 (▪) and _R_t = 8 (•), and (b) _R_t = 12 through an open channel. Notice the difference of timescales in Fig. 8, a and b.
Similar articles
- Floppy but not sloppy: Interaction mechanism of FG-nucleoporins and nuclear transport receptors.
Aramburu IV, Lemke EA. Aramburu IV, et al. Semin Cell Dev Biol. 2017 Aug;68:34-41. doi: 10.1016/j.semcdb.2017.06.026. Epub 2017 Jun 30. Semin Cell Dev Biol. 2017. PMID: 28669824 Free PMC article. Review. - The mechanism of nucleocytoplasmic transport through the nuclear pore complex.
Tetenbaum-Novatt J, Rout MP. Tetenbaum-Novatt J, et al. Cold Spring Harb Symp Quant Biol. 2010;75:567-84. doi: 10.1101/sqb.2010.75.033. Epub 2011 Mar 29. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21447814 - Biomechanics of the transport barrier in the nuclear pore complex.
Stanley GJ, Fassati A, Hoogenboom BW. Stanley GJ, et al. Semin Cell Dev Biol. 2017 Aug;68:42-51. doi: 10.1016/j.semcdb.2017.05.007. Epub 2017 May 12. Semin Cell Dev Biol. 2017. PMID: 28506890 Review. - The selective permeability barrier in the nuclear pore complex.
Li C, Goryaynov A, Yang W. Li C, et al. Nucleus. 2016 Sep 2;7(5):430-446. doi: 10.1080/19491034.2016.1238997. Epub 2016 Sep 27. Nucleus. 2016. PMID: 27673359 Free PMC article. Review. - The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport.
Wälde S, Kehlenbach RH. Wälde S, et al. Trends Cell Biol. 2010 Aug;20(8):461-9. doi: 10.1016/j.tcb.2010.05.001. Epub 2010 Jun 4. Trends Cell Biol. 2010. PMID: 20627572 Review.
Cited by
- Nucleocytoplasmic transport: a thermodynamic mechanism.
Kopito RB, Elbaum M. Kopito RB, et al. HFSP J. 2009;3(2):130-41. doi: 10.2976/1.3080807. Epub 2009 Mar 18. HFSP J. 2009. PMID: 19794817 Free PMC article. - Polymer translocation through a hairy channel mimicking the inner plug of a nuclear pore complex.
Zhang C, Cheng Z, Lin X, Chu W. Zhang C, et al. Eur Biophys J. 2019 May;48(4):317-327. doi: 10.1007/s00249-019-01356-5. Epub 2019 Mar 29. Eur Biophys J. 2019. PMID: 30927020 - From the trap to the basket: getting to the bottom of the nuclear pore complex.
Lim RY, Aebi U, Stoffler D. Lim RY, et al. Chromosoma. 2006 Feb;115(1):15-26. doi: 10.1007/s00412-005-0037-1. Epub 2006 Jan 10. Chromosoma. 2006. PMID: 16402261 - Brownian dynamics simulation of nucleocytoplasmic transport: a coarse-grained model for the functional state of the nuclear pore complex.
Moussavi-Baygi R, Jamali Y, Karimi R, Mofrad MR. Moussavi-Baygi R, et al. PLoS Comput Biol. 2011 Jun;7(6):e1002049. doi: 10.1371/journal.pcbi.1002049. Epub 2011 Jun 2. PLoS Comput Biol. 2011. PMID: 21673865 Free PMC article. - Deciphering the Structure and Function of Nuclear Pores Using Single-Molecule Fluorescence Approaches.
Musser SM, Grünwald D. Musser SM, et al. J Mol Biol. 2016 May 22;428(10 Pt A):2091-119. doi: 10.1016/j.jmb.2016.02.023. Epub 2016 Mar 2. J Mol Biol. 2016. PMID: 26944195 Free PMC article. Review.
References
- Allen, M. P., and D. J. Tildesley. 1987. Computer Simulation of Liquids. Clarendon Press, Oxford, UK.
- Allen, N. P. C., L. Huang, A. Burlingame, and M. Rexach. 2001. Proteonomic analysis of nucleoporin interacting proteins. J. Biol. Chem. 276:29268–29274. - PubMed
- Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear transport. Curr. Opin. Struct. Biol. 11:703–715. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources