Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis - PubMed (original) (raw)
Septal localization of forespore membrane proteins during engulfment in Bacillus subtilis
Aileen Rubio et al. EMBO J. 2004.
Abstract
In Bacillus subtilis, many membrane proteins localize to the sporulation septum, where they play key roles in spore morphogenesis and cell-specific gene expression, but the mechanism for septal targeting is not well understood. SpoIIQ, a forespore-expressed protein, is involved in engulfment and forespore-specific gene expression. We find that SpoIIQ dynamically localizes to the sporulation septum, tracks the engulfing mother cell membrane, assembles into helical arcs around the forespore and is finally degraded. Retention of SpoIIQ in the septum requires one or more mother cell-expressed proteins. We also observed that any forespore-expressed membrane protein initially localizes to the septum and later spreads throughout the forespore membrane, suggesting that membrane protein insertion occurs at the forespore septal region. This possibility provides an attractive mechanism for how activation of mother cell-specific gene expression is restricted to adjacent sister cells, since direct insertion of the signaling protein SpoIIR into the septum would spatially restrict its activity. In keeping with this hypothesis, we find that SpoIIR localizes to the septum and is transiently expressed.
Figures
Figure 1
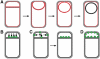
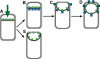
Engulfment pathway of sporulation in B. subtilis and models for targeting of proteins synthesized in the forespore. (A) Cartoon showing B. subtilis engulfment. Following polar septation, the membrane of the larger mother cell migrates around the forespore, until it is completely enclosed. Membranes stained by FM 4-64 are shown in red; following membrane fusion, FM 4-64 no longer stains the forespore membranes, providing an assay for the completion of engulfment. (B–D) Models for targeting forespore-expressed membrane proteins. Proteins (green) can either be directly inserted into the forespore septal region (B) or inserted elsewhere and migrate to the septal region (C). Then, they can either be retained during engulfment (C) or freely diffuse throughout the forespore membrane (D).
Figure 2
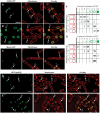
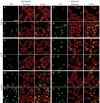
Localization patterns of forespore-expressed heterologous membrane proteins and the native forespore protein SpoIIQ. (A, B) MalF-GFP (green) 2 h (A, hereafter _t_2) and 3 h (B, hereafter _t_3) after the onset of sporulation stained with FM 4-64 (red). In most sporangia, engulfment is underway at _t_2 and complete by _t_3. (C) MotA-GFP (green) at _t_2 stained with Mitotracker Red (red). In panels A and C, arrows 1 and 2 represent sporangia with flat or slightly curving polar septa. In panel B, arrows 3 and 4 represent sporangia before and after membrane fusion, respectively. (D) GFP-SpoIIQ (green) at _t_2 and (E) _t_3 stained with FM 4-64 (red). In panels D and E, arrows 1, 2 and 3 represent early sporangia and arrows 4 and 5 represent sporangia late in engulfment. The scale bar in (A, D) is 2 μm. (F, G) Quantitation of GFP localization at various stages of engulfment (a–d). Numbers refer to the percentage of sporangia in each engulfment class with the indicated localization pattern; those in bold represent the predominant class. (F) GFP-SpoIIQ (AR126) localization. Seven subclasses of GFP localization were scored: (i) none; (ii) medial focus; (iii) ring; (iv) tracking mother cell engulfing membrane; (v) freely diffusible; (vi) punctate; (vii) soluble. A total of 504 sporangia were scored from _t_2 and _t_3. (G) MalF-GFP (AR4) localization. Seven subclasses for GFP localization were scored: (i) none; (ii) focus formation; (iii) tracking mother cell engulfing membrane; (iv) enriched at septum but ahead of engulfing membranes; (v) punctate; (vi) freely diffusible; (vii) soluble. A total of 296 sporangia were scored from _t_2 and _t_3.
Figure 3
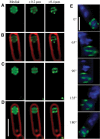
Three-dimensional visualization of GFP-SpoIIQ rings after membrane fusion. Optical sectioning deconvolution microscopy was used to obtain images from different focal planes within the specimen (A–D) and used to reconstruct the three-dimensional model of GFP-SpoIIQ (E). In all, 20 images were obtained from focal planes 0.1 μm (A–D) or 0.15 μm (E) apart. (A–D) Different focal planes of GFP-SpoIIQ (green) from two sporangia, starting with the medial GFP focal plane (far left) and +0.2 and +0.4 μm. For reference, the medial focal plane of FM 4-64-stained membrane (red) is included as an overlay (B, D). (E) Three-dimensional model of GFP-SpoIIQ. Z-stacks each separated by 0.15 μm were loaded into the volume builder function of the Applied Precision SoftWorx program (version 3.3) with modeled projections around the _Y_-axis shown and DAPI-stained chromosomes shown as a reference point (also shown in the Supplementary movie). Scale bar, 2 μm.
Figure 4
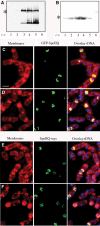
Western blot analysis and immunofluorescence of GFP-SpoIIQ and SpoIIQ-myc. (A) Immunoblot of GFP-SpoIIQ depicting specific proteolytic cleavage during engulfment. Full-length GFP-SpoIIQ is marked with an asterisk (*). (B) Western blot of SpoIIQ-myc shows no smaller degradation product. Full-length SpoIIQ-myc is marked with an asterisk (*). Immunofluorescence samples were taken at (C, E) _t_2 and (D, F) _t_3 and stained with Mitotracker Red (membranes in red), DAPI (DNA in blue) and SpoIIQ (GFP or Myc in green). (C, D) Localization patterns for GFP-SpoIIQ observed for fixed cells are the same as with live cells. (E, F) Localization of SpoIIQ-myc suggests that degradation occurs at the C-terminus of the protein since early patterns are similar to GFP-SpoIIQ, but little signal is observed in late sporangia. Scale bar, 2 μm.
Figure 5
Localization of GFP-SpoIIQ and MalF-GFP in different spo backgrounds. Samples were taken at _t_2 (A, C, E, G, I, K) and _t_3 (B, D, F, H, J, L) after resuspension and stained with FM 4-64 (red, middle column). GFP images (green) are on the left with overlaid images on the right. (A) AR126 (spoIIQ_∷_spc amyE_∷P_spoIIQ gfp-spoIIQ_Ω_cat) at _t_2 and (B) _t_3. (C) AR139 (spoIIQ_∷_spc spoIIP_∷_tet amyE_∷P_spoIIQ gfp-spoIIQ_Ω_cat) at _t_2 and (D) _t_3. GFP-SpoIIQ remains at the septum. (E) AR156 (spoIIQ_∷_spc spoIIGB_∷_erm amyE_∷P_spoIIQ gfp-spoIIQ_Ω_cat) at _t_2 and (F) _t_3. GFP-SpoIIQ diffuses throughout the forespore membrane. (G) AR4 (amyE_∷P_spoIIQ malF 12 -gfp_Ω_cat) at _t_2 and (H) _t_3. (I) AR154 (spoIIP_∷_tet amyE_∷P_spoIIQ malF 12 -gfp_Ω_cat) at _t_2 and (J) _t_3. The arrow in (I) shows enrichment at the septum. (K) AR16 (spoIIGB_∷_erm amyE_∷P_spoIIQ malF 12 -gfp_Ω_cat) at _t_2 and (L) _t_3. Scale bar, 2 μm.
Figure 6
Localization of GFP-SpoIIR. Samples were taken at _t_2 (A) and _t_3 (B) and stained with Mitotracker Red (red). Arrows 1 and 2 represent early sporangia, whereas arrows 3 and 4 represent late sporangia. GFP-SpoIIR (green) first localizes as a focus in the middle of the sporulation septum, and tracks with the engulfing mother cell membrane, but fluorescence is rapidly lost. Scale bar, 2 μm.
Figure 7
Model for localization of forespore membrane proteins. (A) Any membrane protein (green) synthesized by a σF-dependent promoter inserts directly into the forespore septal domain. (B–D) Localized proteins (such as SpoIIQ) are retained through an interaction with a partner protein (blue) made in the mother cell or the septal peptidoglycan. (E) Nonlocalized proteins (such as MalF-GFP) freely diffuse throughout the forespore membrane.
Similar articles
- SpoIIQ anchors membrane proteins on both sides of the sporulation septum in Bacillus subtilis.
Campo N, Marquis KA, Rudner DZ. Campo N, et al. J Biol Chem. 2008 Feb 22;283(8):4975-82. doi: 10.1074/jbc.M708024200. Epub 2007 Dec 11. J Biol Chem. 2008. PMID: 18077456 - Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum.
Doan T, Marquis KA, Rudner DZ. Doan T, et al. Mol Microbiol. 2005 Mar;55(6):1767-81. doi: 10.1111/j.1365-2958.2005.04501.x. Mol Microbiol. 2005. PMID: 15752199 - SpoIIQ-dependent localization of SpoIIE contributes to septal stability and compartmentalization during the engulfment stage of Bacillus subtilis sporulation.
Dehghani B, Rodrigues CDA. Dehghani B, et al. J Bacteriol. 2024 Jul 25;206(7):e0022024. doi: 10.1128/jb.00220-24. Epub 2024 Jun 21. J Bacteriol. 2024. PMID: 38904397 Free PMC article. - Differentiation and the establishment of cell type during sporulation in Bacillus subtilis.
Margolis P, Driks A, Losick R. Margolis P, et al. Curr Opin Genet Dev. 1991 Oct;1(3):330-5. doi: 10.1016/s0959-437x(05)80296-5. Curr Opin Genet Dev. 1991. PMID: 1840889 Review. - Protein Targeting during Bacillus subtilis Sporulation.
Dworkin J. Dworkin J. Microbiol Spectr. 2014 Feb;2(1):TBS-0006-2012. doi: 10.1128/microbiolspec.TBS-0006-2012. Microbiol Spectr. 2014. PMID: 26082125 Review.
Cited by
- Peptidoglycan hydrolysis is required for assembly and activity of the transenvelope secretion complex during sporulation in Bacillus subtilis.
Rodrigues CD, Marquis KA, Meisner J, Rudner DZ. Rodrigues CD, et al. Mol Microbiol. 2013 Sep;89(6):1039-52. doi: 10.1111/mmi.12322. Epub 2013 Aug 1. Mol Microbiol. 2013. PMID: 23834622 Free PMC article. - Engulfment-regulated proteolysis of SpoIIQ: evidence that dual checkpoints control sigma activity.
Jiang X, Rubio A, Chiba S, Pogliano K. Jiang X, et al. Mol Microbiol. 2005 Oct;58(1):102-15. doi: 10.1111/j.1365-2958.2005.04811.x. Mol Microbiol. 2005. PMID: 16164552 Free PMC article. - Loss of compartmentalization of σ(E) activity need not prevent formation of spores by Bacillus subtilis.
Chary VK, Xenopoulos P, Eldar A, Piggot PJ. Chary VK, et al. J Bacteriol. 2010 Nov;192(21):5616-24. doi: 10.1128/JB.00572-10. Epub 2010 Aug 27. J Bacteriol. 2010. PMID: 20802044 Free PMC article. - Remodeling of the Enterococcal Cell Envelope during Surface Penetration Promotes Intrinsic Resistance to Stress.
Ramos Y, Sansone S, Hwang SM, Sandoval TA, Zhu M, Zhang G, Cubillos-Ruiz JR, Morales DK. Ramos Y, et al. mBio. 2022 Dec 20;13(6):e0229422. doi: 10.1128/mbio.02294-22. Epub 2022 Nov 10. mBio. 2022. PMID: 36354750 Free PMC article. - Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation.
Fay A, Meyer P, Dworkin J. Fay A, et al. J Mol Biol. 2010 Jun 18;399(4):547-61. doi: 10.1016/j.jmb.2010.04.036. Epub 2010 Apr 24. J Mol Biol. 2010. PMID: 20417640 Free PMC article.
References
- Addinall SG, Holland B (2002) The tubulin ancestor, FtsZ, draughtsman, designer and driving force for bacterial cytokinesis. J Mol Biol 318: 219–236 - PubMed
- Anderson AJ, Green RS, Sturman AJ, Archibald AR (1978) Cell wall assembly in Bacillus subtilis: location of wall material incorporated during pulsed release of phosphate limitation, its accessibility to bacteriophages and concanavalin A, and its susceptibility to turnover. J Bacteriol 136: 886–899 - PMC - PubMed
- Arigoni F, Guerout-Fleury AM, Barak I, Stragier P (1999) The SpoIIE phosphatase, the sporulation septum and the establishment of forespore-specific transcription in Bacillus subtilis: a reassessment. Mol Microbiol 31: 1407–1415 - PubMed
- Ausmees N, Jacobs-Wagner C (2003) Spatial and temporal control of differentiation and cell cycle progression in Caulobacter crescentus. Annu Rev Microbiol 57: 225–247 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases