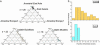Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh - PubMed (original) (raw)
Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh
Thierry Wirth et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The history of mankind remains one of the most challenging fields of study. However, the emergence of anatomically modern humans has been so recent that only a few genetically informative polymorphisms have accumulated. Here, we show that DNA sequences from Helicobacter pylori, a bacterium that colonizes the stomachs of most humans and is usually transmitted within families, can distinguish between closely related human populations and are superior in this respect to classical human genetic markers. H. pylori from Buddhists and Muslims, the two major ethnic communities in Ladakh (India), differ in their population-genetic structure. Moreover, the prokaryotic diversity is consistent with the Buddhists having arisen from an introgression of Tibetan speakers into an ancient Ladakhi population. H. pylori from Muslims contain a much stronger ancestral Ladakhi component, except for several isolates with an Indo-European signature, probably reflecting genetic flux from the Near East. These signatures in H. pylori sequences are congruent with the recent history of population movements in Ladakh, whereas similar signatures in human microsatellites or mtDNA were only marginally significant. H. pylori sequence analysis has the potential to become an important tool for unraveling short-term genetic changes in human populations.
Figures
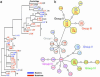
Fig. 1.
Phylogenetic analysis of mtDNA HVS1 sequences. The designations for individual probands consist of L plus an arbitrary number (followed by the haplogroup in a) and are color-coded as indicated. (a) Maximum-likelihood tree of the HVS1 sequences (388 bp) using a Tamura–Nei model of evolution (I = 0.4806; G = 0.7551; starting tree = neighbor-joining; nearest-neighbor interchange branch swapping limited to 20,000 rearrangements). Data were constructed by using
paup
* and bootstrapped with 1,000 replicates. Only bootstrap values >70% are shown. The tree includes the Cambridge Reference Sequence and was rooted with a pygmy sequence. (b) Phylogenetic network relating HVS1 sequences. Colors of the filled circles correspond to the six main radiation groups described in Oota et al. (38), and small open circles correspond to missing haplotypes. The network obtained from Ladakhi HVS1 sequences is completely congruent with the network of Oota et al. (38), except that proband L133 shares typical Group II variant sites (C at np362 and G at np319) but is placed at a terminal node of Group VI.
Fig. 2.
Three-dimensional correspondence analysis (FCA) of microsatellite genotypes from Ladakhis. Yellow, Buddhists; blue, Muslims.
Fig. 3.
A multilocus haplotype tree and quantitative sources of nucleotides from three ancestral populations for 119 H. pylori isolates from Ladakh, East Asia, and Indo-Europe. (a) Neighbor-joining tree of individual haplotypes (Kimura two-parameter model). Fig. 6, which is published as supporting information on the PNAS web site, includes the designation of each individual isolate. (b) Sources of nucleotides. The diameters of the three circles indicate the genetic diversity of the three ancestral populations and the filled arcs indicate the proportion of the nucleotides in the 119 haplotypes that are derived from each of these populations. The black lines joining the ancestral populations represent a neighbor-joining population tree as measured by , the net nucleotide distance between populations.
Fig. 4.
Ancestry of nucleotides in H. pylori isolates from Ladakh as inferred by
structure
.(a) Ternary plots of proportion of ancestry from three sources of nucleotides designated ancestral East Asia, ancestral Europe1, and ancestral Europe2. Each data point corresponds to a single isolate whose proportion of ancestry from each of the three sources is represented by its proximity to the corresponding corner of the triangle. Note that the proportion of ancestry ranges between 0 and 1.0, regardless of the true genetic distances between the ancestral sources. (b) Proportion of ancestry from ancestral East Asia for H. pylori isolates by ethnic source.
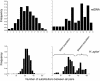
Fig. 5.
Mismatch distributions of differences between all possible pairs of HVS1 and H. pylori sequences from Buddhists (Left) and Muslims (Right). The assignment of data in the lower right to within- and between-population components is based on the observation that the between-population component disappeared when the three isolates with considerable ancestry from Europe2 were excluded.
Similar articles
- Helicobacter pylori Sequences Reflect Past Human Migrations.
Moodley Y, Linz B. Moodley Y, et al. Genome Dyn. 2009;6:62-74. doi: 10.1159/000235763. Epub 2009 Aug 19. Genome Dyn. 2009. PMID: 19696494 - Ethnic and geographic differentiation of Helicobacter pylori within Iran.
Latifi-Navid S, Ghorashi SA, Siavoshi F, Linz B, Massarrat S, Khegay T, Salmanian AH, Shayesteh AA, Masoodi M, Ghanadi K, Ganji A, Suerbaum S, Achtman M, Malekzadeh R, Falush D. Latifi-Navid S, et al. PLoS One. 2010 Mar 22;5(3):e9645. doi: 10.1371/journal.pone.0009645. PLoS One. 2010. PMID: 20339588 Free PMC article. - Impact of human migrations on diversity of Helicobacter pylori in Cambodia and New Caledonia.
Breurec S, Raymond J, Thiberge JM, Hem S, Monchy D, Seck A, Dehoux P, Garin B, Dauga C. Breurec S, et al. Helicobacter. 2013 Aug;18(4):249-61. doi: 10.1111/hel.12037. Epub 2013 Jan 28. Helicobacter. 2013. PMID: 23350664 - Discovering human history from stomach bacteria.
Disotell TR. Disotell TR. Genome Biol. 2003;4(5):213. doi: 10.1186/gb-2003-4-5-213. Epub 2003 Apr 28. Genome Biol. 2003. PMID: 12734001 Free PMC article. Review. - Deciphering host migrations and origins by means of their microbes.
Wirth T, Meyer A, Achtman M. Wirth T, et al. Mol Ecol. 2005 Oct;14(11):3289-306. doi: 10.1111/j.1365-294X.2005.02687.x. Mol Ecol. 2005. PMID: 16156803 Review.
Cited by
- The human microbiome: at the interface of health and disease.
Cho I, Blaser MJ. Cho I, et al. Nat Rev Genet. 2012 Mar 13;13(4):260-70. doi: 10.1038/nrg3182. Nat Rev Genet. 2012. PMID: 22411464 Free PMC article. Review. - An African origin for the intimate association between humans and Helicobacter pylori.
Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. Linz B, et al. Nature. 2007 Feb 22;445(7130):915-918. doi: 10.1038/nature05562. Epub 2007 Feb 7. Nature. 2007. PMID: 17287725 Free PMC article. - Recent acquisition of Helicobacter pylori by Baka pygmies.
Nell S, Eibach D, Montano V, Maady A, Nkwescheu A, Siri J, Elamin WF, Falush D, Linz B, Achtman M, Moodley Y, Suerbaum S. Nell S, et al. PLoS Genet. 2013;9(9):e1003775. doi: 10.1371/journal.pgen.1003775. Epub 2013 Sep 19. PLoS Genet. 2013. PMID: 24068950 Free PMC article. - Evolutionary history of Helicobacter pylori sequences reflect past human migrations in Southeast Asia.
Breurec S, Guillard B, Hem S, Brisse S, Dieye FB, Huerre M, Oung C, Raymond J, Tan TS, Thiberge JM, Vong S, Monchy D, Linz B. Breurec S, et al. PLoS One. 2011;6(7):e22058. doi: 10.1371/journal.pone.0022058. Epub 2011 Jul 19. PLoS One. 2011. PMID: 21818291 Free PMC article. - Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer.
McClain MS, Shaffer CL, Israel DA, Peek RM Jr, Cover TL. McClain MS, et al. BMC Genomics. 2009 Jan 5;10:3. doi: 10.1186/1471-2164-10-3. BMC Genomics. 2009. PMID: 19123947 Free PMC article.
References
- Cann, R. L. (2001) Science 291, 1742–1748. - PubMed
- Cavalli-Sforza, L. L. (1998) TIG 14, 60–65. - PubMed
- Goldstein, D. B. & Chikhi, L. (2002) Annu. Rev. Genomics Hum. Genet. 3, 129–152. - PubMed
- Cann, R. L., Stoneking, M. & Wilson, A. C. (1987) Nature 325, 31–36. - PubMed
- Cavalli-Sforza, L. L. & Feldman, M. W. (2003) Nat. Genet. 33, Suppl., 266–275. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials