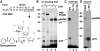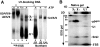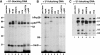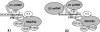p54(nrb) associates with the 5' splice site within large transcription/splicing complexes - PubMed (original) (raw)
p54(nrb) associates with the 5' splice site within large transcription/splicing complexes
Sei Kameoka et al. EMBO J. 2004.
Abstract
The functional coupling of transcription and splicing has been reported both in vivo and in vitro, but the molecular mechanisms governing these interactions remain largely unknown. Here we show that p54(nrb), a transcription/splicing factor, associates with the 5' splice site (SS) within large complexes present in HeLa cell nuclear extracts, in which the hyperphosphorylated form of RNA polymerase II (RNAPIIO) is associated with U1 or U1 and U2 snRNPs. These RNAPIIO-snRNP complexes also contain other transcription/splicing factors, such as PSF and TLS, as well as transcription factors that interact with RNAPIIO during elongation, including P-TEFb, TAT-SF1 and TFIIF. The presence of these factors in functional elongation complexes, demonstrated using an immobilized DNA template assay, strongly suggests that the RNAPIIO-snRNP complexes reflect physiologically relevant interactions between the transcription and splicing machineries. Our finding that both p54(nrb) and PSF, which bind the C-terminal domain of the largest subunit of RNAPII, can interact directly with the 5' SS indicates that these factors may mediate contacts between RNAPII and snRNPs during the coupled transcription/splicing process.
Figures
Figure 1
Crosslinking profile of the BP-derivatized 5′SS RNA. (A) Structure of the 32P-labeled, BP-derivatized 14-nt 5′SS RNA. (B) The 5′SS RNA was incubated with HeLa NE in the absence (lanes 1, 3) or presence (lanes 2, 4) of ATP and in the presence (lanes 1, 2) or absence (lanes 3, 4) of U1-blocking DNA. (C) Wild-type 5′SS RNA (WT) or 5′SS RNAs with single point mutations at intron pos. +1 or +2 was incubated with HeLa NE in the absence of ATP or U1-blocking DNA. (D) The 5′SS crosslinks were immunoprecipitated under denaturing conditions with α-Sm (Y12) antibodies. Crosslinked products were resolved in 11% SDS gels. Positions of the molecular weight markers and the 5′SS crosslinks are indicated.
Figure 2
The 5′SS:p54/56 crosslink corresponds to p54nrb. (A) The 5′SS:p54/56 crosslinks were treated with RNase A (lane 2) or RNase A and phosphatase (CIP) (lane 3) and resolved in an 8% SDS gel. (B) The 5′SS crosslinks were immunoprecipitated under denaturing (lanes 1–4) or native (lanes 5 and 6) conditions using the indicated antibodies and resolved in an 11% SDS gel. (C) The 5′SS:p54nrb and 5′SS:hPrp28 crosslinks were purified from an SDS gel, mixed (lane 1), immunoprecipitated with pre-immune serum (lane 2) or the indicated antibodies (lanes 3 and 4) and resolved in an 8% SDS gel. (D) Diagram of the human PSF and p54nrb sequences. The two RRMs, the proline/glutamine-rich regions (PQ), the 293-aa internal region of high similarity between p54nrb and PSF, the sequences of three Lys-C peptides and the mapped region of the 5′SS crosslink within p54nrb (aa 18–53) are indicated.
Figure 3
The 5′SS:p54nrb crosslink exists exclusively in large complexes containing U1 snRNP. (A) Standard binding reactions in the presence (lanes 1–4) or absence (lanes 5–7) of the 5′SS RNA, in the absence (lanes 1,3) or presence (lanes 2,4–7) of ATP and in the presence (lanes 1,2) or absence (lanes 3–7) of U1-blocking DNA were resolved in a native 4% gel. snRNP complexes were detected by autoradiography (lanes 1–4) or Northern hybridization with U1, U2 or U4 probes (lanes 5–7). Positions of complexes are indicated. (B) The 5′SS crosslinks were resolved in a native 4% gel in the first dimension and in an 11% SDS gel in the second. Positions of the free 5′SS and the crosslinks are indicated.
Figure 4
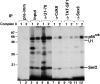
X1 and X2 complexes can be separated in a glycerol gradient. A 150-μl crosslinking reaction performed in the presence of ATP and absence of U1-blocking DNA was separated in a 10–30% glycerol gradient. In all, 10 μl of each fraction was resolved in either a 4% native gel (A) or an 11% SDS gel (B). (C) Fractions 4 and 7 of the glycerol gradient were immunoprecipitated under native conditions using the indicated antibodies, and the bound material was resolved in an 11% SDS gel. The identity of the ∼70 kDa signal generated with the α-TLS antibody (lane 21) has not been determined. Positions of the 5′SS crosslinks are indicated.
Figure 5
The p54nrb crosslink in X1 and X2 complexes is associated with RNAPIIO. A crosslinking reaction performed in the presence of ATP and in the absence (A) or presence (B) of U1-blocking DNA was immunoprecipitated under native conditions using the indicated α-RNAPII antibodies and the bound material resolved in an 11% SDS gel. The identity of the ∼65 kDa signal (e.g. lane 3 in panel A) has not been determined. (C) HeLa NE was incubated with ATP in the absence of the 5′SS RNA and immunoprecipitated as in (A). The bound RNA was analyzed by Northern hybridization with a mixture of U1, U2, U4, U5 and U6 snRNA probes. Positions of snRNAs are indicated. Although co-migrating with U6, the signal indicated by an arrow in lane 4 represents a degradation product of U1 snRNA.
Figure 6
X1 and X2 complexes contain several transcription elongation factors. A crosslinking reaction performed in the presence of ATP was separated in a glycerol gradient, and fractions containing X1 (fraction 4) and X2 (fraction 7) complexes were immunoprecipitated under native conditions using the indicated antibodies and the bound material was resolved in an 11% SDS gel.
Figure 7
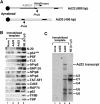
X1 and X2 complexes contain a subset of factors present in functional transcription ECs. (A) Schematic representation of the immobilized Ad20 and Ad22 DNA templates, showing the AdMLP, the two leading exons of the adenovirus major late transcription unit (black boxes), the start site for RNAPII transcription and the unique _Pvu_II restriction sites. (B) Western blot analysis of protein factors bound to template fragments released from beads lacking DNA or linked to the Ad20 or Ad22 templates using the indicated antibodies. (C) Northern blot analysis of template fragments released from beads lacking DNA or linked to the Ad20 or Ad22 templates using probes for all the five U snRNAs and the Ad22 transcript.
Figure 8
Schematic composition of X1 and X2 complexes. Relative positions of the indicated components are arbitrary.
Similar articles
- Structure, Dynamics, and Interaction of p54(nrb)/NonO RRM1 with 5' Splice Site RNA Sequence.
Duvignaud JB, Bédard M, Nagata T, Muto Y, Yokoyama S, Gagné SM, Vincent M. Duvignaud JB, et al. Biochemistry. 2016 May 10;55(18):2553-66. doi: 10.1021/acs.biochem.5b01240. Epub 2016 Apr 26. Biochemistry. 2016. PMID: 27064654 - Brm transactivates the telomerase reverse transcriptase (TERT) gene and modulates the splicing patterns of its transcripts in concert with p54(nrb).
Ito T, Watanabe H, Yamamichi N, Kondo S, Tando T, Haraguchi T, Mizutani T, Sakurai K, Fujita S, Izumi T, Isobe T, Iba H. Ito T, et al. Biochem J. 2008 Apr 1;411(1):201-9. doi: 10.1042/BJ20071075. Biochem J. 2008. PMID: 18042045 - The mitotic phosphorylation of p54(nrb) modulates its RNA binding activity.
Bruelle C, Bédard M, Blier S, Gauthier M, Traish AM, Vincent M. Bruelle C, et al. Biochem Cell Biol. 2011 Aug;89(4):423-33. doi: 10.1139/o11-030. Biochem Cell Biol. 2011. PMID: 21819346 - Ser/Arg-rich protein-mediated communication between U1 and U2 small nuclear ribonucleoprotein particles.
Boukis LA, Liu N, Furuyama S, Bruzik JP. Boukis LA, et al. J Biol Chem. 2004 Jul 9;279(28):29647-53. doi: 10.1074/jbc.M313209200. Epub 2004 May 5. J Biol Chem. 2004. PMID: 15131126 - How Are Short Exons Flanked by Long Introns Defined and Committed to Splicing?
Hollander D, Naftelberg S, Lev-Maor G, Kornblihtt AR, Ast G. Hollander D, et al. Trends Genet. 2016 Oct;32(10):596-606. doi: 10.1016/j.tig.2016.07.003. Epub 2016 Aug 6. Trends Genet. 2016. PMID: 27507607 Review.
Cited by
- The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration.
Lim YW, James D, Huang J, Lee M. Lim YW, et al. Int J Mol Sci. 2020 Sep 28;21(19):7151. doi: 10.3390/ijms21197151. Int J Mol Sci. 2020. PMID: 32998269 Free PMC article. Review. - Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA.
Chen LL, Carmichael GG. Chen LL, et al. Mol Cell. 2009 Aug 28;35(4):467-78. doi: 10.1016/j.molcel.2009.06.027. Mol Cell. 2009. PMID: 19716791 Free PMC article. - Pathogenesis of Frontotemporal Lobar Degeneration: Insights From Loss of Function Theory and Early Involvement of the Caudate Nucleus.
Sobue G, Ishigaki S, Watanabe H. Sobue G, et al. Front Neurosci. 2018 Jul 12;12:473. doi: 10.3389/fnins.2018.00473. eCollection 2018. Front Neurosci. 2018. PMID: 30050404 Free PMC article. Review. - Compromised paraspeckle formation as a pathogenic factor in FUSopathies.
Shelkovnikova TA, Robinson HK, Troakes C, Ninkina N, Buchman VL. Shelkovnikova TA, et al. Hum Mol Genet. 2014 May 1;23(9):2298-312. doi: 10.1093/hmg/ddt622. Epub 2013 Dec 11. Hum Mol Genet. 2014. PMID: 24334610 Free PMC article. - The transcribed ultraconserved element uc.51 promotes the proliferation and metastasis of breast cancer by stabilizing NONO.
Shi X, Huang X, Chen R, Li Y, Xu Y, Zhang W, Zhu Q, Zha X, Wang J. Shi X, et al. Clin Exp Metastasis. 2021 Dec;38(6):551-571. doi: 10.1007/s10585-021-10128-5. Epub 2021 Oct 29. Clin Exp Metastasis. 2021. PMID: 34714466
References
- Bauren G, Wieslander L (1994) Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell 76: 183–192 - PubMed
- Bentley D (2002) The mRNA assembly line: transcription and processing machines in the same factory. Curr Opin Cell Biol 14: 336–342 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources