Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia - PubMed (original) (raw)
. 2004 Apr;113(7):1008-16.
doi: 10.1172/JCI19399.
Franco Fais, Angelo Valetto, Emilia Albesiano, Shiori Hashimoto, Mariella Dono, Hideyuki Ikematsu, Steven L Allen, Jonathan Kolitz, Kanti R Rai, Marco Nardini, Anna Tramontano, Manlio Ferrarini, Nicholas Chiorazzi
Affiliations
- PMID: 15057307
- PMCID: PMC379317
- DOI: 10.1172/JCI19399
Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia
Fabio Ghiotto et al. J Clin Invest. 2004 Apr.
Abstract
Studies of B cell antigen receptors (BCRs) expressed by leukemic lymphocytes from patients with B cell chronic lymphocytic leukemia (B-CLL) suggest that B lymphocytes with some level of BCR structural restriction become transformed. While analyzing rearranged V(H)DJ(H) and V(L)J(L) genes of 25 non-IgM-producing B-CLL cases, we found five IgG(+) cases that display strikingly similar BCRs (use of the same H- and L-chain V gene segments with unique, shared heavy chain third complementarity-determining region [HCDR3] and light chain third complementarity-determining region [LCDR3] motifs). These H- and L-chain characteristics were not identified in other B-CLL cases or in normal B lymphocytes whose sequences are available in the public databases. Three-dimensional modeling studies suggest that these BCRs could bind the same antigenic epitope. The structural features of the B-CLL BCRs resemble those of mAb's reactive with carbohydrate determinants of bacterial capsules or viral coats and with certain autoantigens. These findings suggest that the B lymphocytes that gave rise to these IgG(+) B-CLL cells were selected for this unique BCR structure. This selection could have occurred because the precursors of the B-CLL cells were chosen for their antigen-binding capabilities by antigen(s) of restricted nature and structure, or because the precursors derived from a B cell subpopulation with limited BCR heterogeneity, or both.
Figures
Figure 1
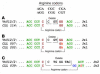
Amino acid sequences of HCDR3 and LCDR3 of IgG+ B-CLL cases. Amino acid sequences are shown flanked by the 3′ end of FR3 and the 5′ end of FR4. A consensus sequence is shown for each. Color code: identical amino acids are in brown, chemically similar amino acids in blue, unrelated amino acids in black, and differences within areas of identity or similarity in red. (A) HCDR3. The consensus sequence consists of two hydrophilic amino acids (blue), seven amino acids that represent a portion of the germline D6–13 gene segment (brown), a glycine or serine (both with small side chains; brown), one to two variable amino acids, and five amino acids from the 5′ portion of the germline JH5b segment. (B) LCDR3. The LCDR3 sequences of all cases are identical, except for a difference in the last amino acid in CLL no. 039, due to the use of the Jκ2 segment. In each case, an arginine occurs at the Vκ-Jκ junction (underlined and in italics).
Figure 2
Different mechanisms generate an arginine at the VL-JL junctions. The codons for arginine are listed. Color code: identical nucleotides of the arginine codon are in brown, nucleotides deleted are in black, nucleotides probably resulting from nontemplated nucleotide addition are in dark blue, and differences within areas of identity or similarity in red. (A and B) An appropriate codon results from coding end trimming and recombination of the two germline Vκ and Jκ1 segments. (C) The arginine codon develops via a more complex combinatorial process, requiring the trimming of one nucleotide from the 3′ end of VL, the deletion of seven nucleotides from the 5′ end of Jκ2, and the nontemplated insertion of two G nucleotides.
Figure 3
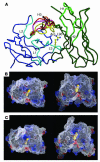
C-alpha trace of the structural models produced by WAM. (A) Overlay of main chains of the five CLL three-dimensional structures. For CLL no. 209, the consensus L-chain amino acid sequence was added for WAM analysis. B-CLL L chains are shown in blue color (hypervariable loops L1, L2, and L3 in light blue), and H chains are shown in green color (hypervariable loops H1, and H2 in light green). The different H3 loop structures are color-coded, as noted by the lime green, red, black, purple, and dark green lines in the H3 area. Figure was prepared with MOLSCRIPT (91), and Raster3D (92). (B and C) Electrostatic and molecular surface representation of the structure of CLL no. 039 as a model representative of the B-CLL cases of nonbulged conformation (nos. 039 and 114; B) and of the CLL no. 057 as a model of the cases of bulged conformation (nos. 057, 202, and 209; C). Negative surface potentials are indicated in red, positive surface potentials in blue, and neutral potentials in white. The arginine L96 side chain is shown in yellow CPK representation. The structures are shown from a top view (left panels) and from a side view, corresponding to an approximately 70° rotation relative to the top view (right panels). Figures were prepared with GRASP (93). Arg L96; arginine L96 side chain.
Comment in
- Ig V region restrictions in human chronic lymphocytic leukemia suggest some cases have a common origin.
Kolar GR, Capra JD. Kolar GR, et al. J Clin Invest. 2004 Apr;113(7):952-4. doi: 10.1172/JCI21412. J Clin Invest. 2004. PMID: 15057298 Free PMC article.
Similar articles
- B cell receptors in TCL1 transgenic mice resemble those of aggressive, treatment-resistant human chronic lymphocytic leukemia.
Yan XJ, Albesiano E, Zanesi N, Yancopoulos S, Sawyer A, Romano E, Petlickovski A, Efremov DG, Croce CM, Chiorazzi N. Yan XJ, et al. Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11713-8. doi: 10.1073/pnas.0604564103. Epub 2006 Jul 24. Proc Natl Acad Sci U S A. 2006. PMID: 16864779 Free PMC article. - B-cell chronic lymphocytic leukemia, a clonal disease of B lymphocytes with receptors that vary in specificity for (auto)antigens.
Chiorazzi N, Hatzi K, Albesiano E. Chiorazzi N, et al. Ann N Y Acad Sci. 2005 Dec;1062:1-12. doi: 10.1196/annals.1358.002. Ann N Y Acad Sci. 2005. PMID: 16461783 Review. - Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia.
Tobin G, Thunberg U, Karlsson K, Murray F, Laurell A, Willander K, Enblad G, Merup M, Vilpo J, Juliusson G, Sundström C, Söderberg O, Roos G, Rosenquist R. Tobin G, et al. Blood. 2004 Nov 1;104(9):2879-85. doi: 10.1182/blood-2004-01-0132. Epub 2004 Jun 24. Blood. 2004. PMID: 15217826 - Chronic lymphocytic leukemia: a tale of one or two signals?
Chiorazzi N, Efremov DG. Chiorazzi N, et al. Cell Res. 2013 Feb;23(2):182-5. doi: 10.1038/cr.2012.152. Epub 2012 Nov 13. Cell Res. 2013. PMID: 23147791 Free PMC article. - What is the current evidence for antigen involvement in the development of chronic lymphocytic leukemia?
Tobin G, Rosén A, Rosenquist R. Tobin G, et al. Hematol Oncol. 2006 Mar;24(1):7-13. doi: 10.1002/hon.760. Hematol Oncol. 2006. PMID: 16315334 Review.
Cited by
- A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi.
Hoogeboom R, van Kessel KP, Hochstenbach F, Wormhoudt TA, Reinten RJ, Wagner K, Kater AP, Guikema JE, Bende RJ, van Noesel CJ. Hoogeboom R, et al. J Exp Med. 2013 Jan 14;210(1):59-70. doi: 10.1084/jem.20121801. Epub 2013 Jan 7. J Exp Med. 2013. PMID: 23296468 Free PMC article. - Intrinsic and extrinsic factors influencing the clinical course of B-cell chronic lymphocytic leukemia: prognostic markers with pathogenetic relevance.
Dal-Bo M, Bertoni F, Forconi F, Zucchetto A, Bomben R, Marasca R, Deaglio S, Laurenti L, Efremov DG, Gaidano G, Del Poeta G, Gattei V. Dal-Bo M, et al. J Transl Med. 2009 Aug 28;7:76. doi: 10.1186/1479-5876-7-76. J Transl Med. 2009. PMID: 19715592 Free PMC article. Review. - Distinctive Signaling Profiles With Distinct Biological and Clinical Implications in Aggressive CLL Subsets With Stereotyped B-Cell Receptor Immunoglobulin.
Gerousi M, Laidou S, Gemenetzi K, Stamatopoulos K, Chatzidimitriou A. Gerousi M, et al. Front Oncol. 2021 Nov 3;11:771454. doi: 10.3389/fonc.2021.771454. eCollection 2021. Front Oncol. 2021. PMID: 34804974 Free PMC article. Review. - B cell receptor isotypes differentially associate with cell signaling, kinetics, and outcome in chronic lymphocytic leukemia.
Mazzarello AN, Gentner-Göbel E, Dühren-von Minden M, Tarasenko TN, Nicolò A, Ferrer G, Vergani S, Liu Y, Bagnara D, Rai KR, Burger JA, McGuire PJ, Maity PC, Jumaa H, Chiorazzi N. Mazzarello AN, et al. J Clin Invest. 2022 Jan 18;132(2):e149308. doi: 10.1172/JCI149308. J Clin Invest. 2022. PMID: 34813501 Free PMC article. - Differential organization of tonic and chronic B cell antigen receptors in the plasma membrane.
Gomes de Castro MA, Wildhagen H, Sograte-Idrissi S, Hitzing C, Binder M, Trepel M, Engels N, Opazo F. Gomes de Castro MA, et al. Nat Commun. 2019 Feb 18;10(1):820. doi: 10.1038/s41467-019-08677-1. Nat Commun. 2019. PMID: 30778055 Free PMC article.
References
- Rai, K.R., and Patel, D.V. 1995. Chronic lymphocytic leukemia. In Hematology: basic principles and practice. R. Hoffman, E. Benz, S. Shattil, B. Furie, H. Cohen, and L. Silberstein, editors. Churchill Livingstone, New York. 1308–1321.
- Rozman C, Montserrat E. Chronic lymphocytic leukemia. N. Engl. J. Med. 1995;333:1052–1057. - PubMed
- Oscier DG. Cytogenetic and molecular abnormalities in chronic lymphocytic leukaemia. Blood Rev. 1994;8:88–97. - PubMed
- Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Ann. Rev. Immunol. 2003;21:841–894. - PubMed
- Valetto A, et al. A subset of IgG+ B-CLL cells expresses virtually identical antigens receptors that bind similar peptides. Blood. 1998;92:431a.
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA087956/CA/NCI NIH HHS/United States
- R01 CA81554/CA/NCI NIH HHS/United States
- M01 RR018535/RR/NCRR NIH HHS/United States
- R01 CA081554/CA/NCI NIH HHS/United States
- R01 CA 87956/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources