Recent segmental duplications in the working draft assembly of the brown Norway rat - PubMed (original) (raw)
Recent segmental duplications in the working draft assembly of the brown Norway rat
Eray Tuzun et al. Genome Res. 2004 Apr.
Abstract
We assessed the content, structure, and distribution of segmental duplications (> or =90% sequence identity, > or =5 kb length) within the published version of the Rattus norvegicus genome assembly (v.3.1). The overall fraction of duplicated sequence within the rat assembly (2.92%) is greater than that of the mouse (1%-1.2%) but significantly less than that of human ( approximately 5%). Duplications were nonuniformly distributed, occurring predominantly as tandem and tightly clustered intrachromosomal duplications. Regions containing extensive interchromosomal duplications were observed, particularly within subtelomeric and pericentromeric regions. We identified 41 discrete genomic regions greater than 1 Mb in size, termed "duplication blocks." These appear to have been the target of extensive duplication over millions of years of evolution. Gene content within duplicated regions ( approximately 1%) was lower than expected based on the genome representation. Interestingly, sequence contigs lacking chromosome assignment ("the unplaced chromosome") showed a marked enrichment for segmental duplication (45% of 75.2 Mb), indicating that segmental duplications have been problematic for sequence and assembly of the rat genome. Further targeted efforts are required to resolve the organization and complexity of these regions.
Figures
Figure 1
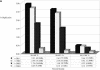
Duplicated fraction in the rat genome. The figure depicts the proportion of the genome that shows duplication (A) when all genomic sequence was compared, and (B) for the rat genome excluding random, unassigned sequence contigs. Various lengths and % identity thresholds are shown. A very small portion of the rat genome shows segmental duplications with ≥99.5% sequence identity. This suggests that the majority of segmental duplications are bona fide and are not the result of missed allelic overlaps during genome assembly.
Figure 1
Duplicated fraction in the rat genome. The figure depicts the proportion of the genome that shows duplication (A) when all genomic sequence was compared, and (B) for the rat genome excluding random, unassigned sequence contigs. Various lengths and % identity thresholds are shown. A very small portion of the rat genome shows segmental duplications with ≥99.5% sequence identity. This suggests that the majority of segmental duplications are bona fide and are not the result of missed allelic overlaps during genome assembly.
Figure 2
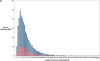
Sequence properties of rat segmental duplications. Distributions of the (A) length and (B) percent nucleotide sequence identity for segmental duplications are shown as a function of the number of aligned bp. Interchromosomal duplications (red); intrachromosomal duplications (blue).
Figure 2
Sequence properties of rat segmental duplications. Distributions of the (A) length and (B) percent nucleotide sequence identity for segmental duplications are shown as a function of the number of aligned bp. Interchromosomal duplications (red); intrachromosomal duplications (blue).
Figure 3
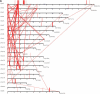
Distribution of segmental duplications (≥90% and ≥10 kb) in the rat genome. The pattern of (A) interchromosomal duplications (red) and (B) intrachromosomal duplications (blue) are depicted for all duplications ≥90% sequence identity and ≥10 kb in length. For clarity, interchromosomal distribution patterns with the random, unassigned sequence contigs (chrUn) are not shown for (A). For more detail, including % identity and pairwise relationships of all duplications and alignments, see http://ratparalogy.cwru.edu.
Figure 3
Distribution of segmental duplications (≥90% and ≥10 kb) in the rat genome. The pattern of (A) interchromosomal duplications (red) and (B) intrachromosomal duplications (blue) are depicted for all duplications ≥90% sequence identity and ≥10 kb in length. For clarity, interchromosomal distribution patterns with the random, unassigned sequence contigs (chrUn) are not shown for (A). For more detail, including % identity and pairwise relationships of all duplications and alignments, see http://ratparalogy.cwru.edu.
Figure 4
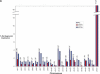
(A) Segmental duplication content per chromosome. The relative proportion of intrachromosomal and interchromosomal duplications for each chromosome is shown. The above calculations treat the unmapped sequence as a separate chromosome when classifying duplications as inter- or intrachromosomal. Forty-five percent of the unplaced chromosome is made up almost entirely of duplicated sequence. (B) Duplication blocks. Rat segmental duplications clustered into larger regions ranging from 100 to 3000 kb in length. We termed these structures “duplication blocks.” Examples of duplication blocks on chromosomes 1 and 7 are presented (arrows) with the underlying degree of sequence identity for each pairwise depicted below the graph. Chromosome 1, green; chromosome 7, red. A subtelomeric (t) and pericentromeric (p) block are indicated. The regions of the rat genome are typified by low gene density (RefSeq/EST/mRNA), a high frequency of gaps within the assembly, and an excess of pairwise alignments.
Figure 4
(A) Segmental duplication content per chromosome. The relative proportion of intrachromosomal and interchromosomal duplications for each chromosome is shown. The above calculations treat the unmapped sequence as a separate chromosome when classifying duplications as inter- or intrachromosomal. Forty-five percent of the unplaced chromosome is made up almost entirely of duplicated sequence. (B) Duplication blocks. Rat segmental duplications clustered into larger regions ranging from 100 to 3000 kb in length. We termed these structures “duplication blocks.” Examples of duplication blocks on chromosomes 1 and 7 are presented (arrows) with the underlying degree of sequence identity for each pairwise depicted below the graph. Chromosome 1, green; chromosome 7, red. A subtelomeric (t) and pericentromeric (p) block are indicated. The regions of the rat genome are typified by low gene density (RefSeq/EST/mRNA), a high frequency of gaps within the assembly, and an excess of pairwise alignments.
Similar articles
- Recent segmental and gene duplications in the mouse genome.
Cheung J, Wilson MD, Zhang J, Khaja R, MacDonald JR, Heng HH, Koop BF, Scherer SW. Cheung J, et al. Genome Biol. 2003;4(8):R47. doi: 10.1186/gb-2003-4-8-r47. Epub 2003 Jul 9. Genome Biol. 2003. PMID: 12914656 Free PMC article. - Segmental duplications: organization and impact within the current human genome project assembly.
Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Bailey JA, et al. Genome Res. 2001 Jun;11(6):1005-17. doi: 10.1101/gr.gr-1871r. Genome Res. 2001. PMID: 11381028 Free PMC article. - Patterns of segmental duplication in the human genome.
Zhang L, Lu HH, Chung WY, Yang J, Li WH. Zhang L, et al. Mol Biol Evol. 2005 Jan;22(1):135-41. doi: 10.1093/molbev/msh262. Epub 2004 Sep 15. Mol Biol Evol. 2005. PMID: 15371527 - Recent duplication, domain accretion and the dynamic mutation of the human genome.
Eichler EE. Eichler EE. Trends Genet. 2001 Nov;17(11):661-9. doi: 10.1016/s0168-9525(01)02492-1. Trends Genet. 2001. PMID: 11672867 Review. - Lessons from the human genome: transitions between euchromatin and heterochromatin.
Horvath JE, Bailey JA, Locke DP, Eichler EE. Horvath JE, et al. Hum Mol Genet. 2001 Oct 1;10(20):2215-23. doi: 10.1093/hmg/10.20.2215. Hum Mol Genet. 2001. PMID: 11673404 Review.
Cited by
- Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome.
Zhang Z, Burch PE, Cooney AJ, Lanz RB, Pereira FA, Wu J, Gibbs RA, Weinstock G, Wheeler DA. Zhang Z, et al. Genome Res. 2004 Apr;14(4):580-90. doi: 10.1101/gr.2160004. Genome Res. 2004. PMID: 15059999 Free PMC article. - A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications.
She X, Liu G, Ventura M, Zhao S, Misceo D, Roberto R, Cardone MF, Rocchi M; NISC Comparative Sequencing Program; Green ED, Archidiacano N, Eichler EE. She X, et al. Genome Res. 2006 May;16(5):576-83. doi: 10.1101/gr.4949406. Epub 2006 Apr 10. Genome Res. 2006. PMID: 16606706 Free PMC article. - Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization.
Goidts V, Armengol L, Schempp W, Conroy J, Nowak N, Müller S, Cooper DN, Estivill X, Enard W, Szamalek JM, Hameister H, Kehrer-Sawatzki H. Goidts V, et al. Hum Genet. 2006 Mar;119(1-2):185-98. doi: 10.1007/s00439-005-0130-9. Epub 2006 Jan 5. Hum Genet. 2006. PMID: 16395594 - Analysis of recent segmental duplications in the bovine genome.
Liu GE, Ventura M, Cellamare A, Chen L, Cheng Z, Zhu B, Li C, Song J, Eichler EE. Liu GE, et al. BMC Genomics. 2009 Dec 1;10:571. doi: 10.1186/1471-2164-10-571. BMC Genomics. 2009. PMID: 19951423 Free PMC article. - Dynamic building of a BAC clone tiling path for the Rat Genome Sequencing Project.
Chen R, Sodergren E, Weinstock GM, Gibbs RA. Chen R, et al. Genome Res. 2004 Apr;14(4):679-84. doi: 10.1101/gr.2171704. Genome Res. 2004. PMID: 15060010 Free PMC article.
References
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. - PubMed
WEB SITE REFERENCES
- http://ratparalogy.cwru.edu; Segmental Duplication Database for Rat at CWRU.
- http://genome.ucsc.edu; Genome browser at Univ. California–Santa Cruz.
- http://www.hgsc.bcm.tmc.edu/; Human Genome Sequencing Center at Baylor College of Medicine. - PubMed
- http://rgd.mcw.edu/; Rat Genome Database at Medical College of Wisconsin.
Publication types
MeSH terms
Grants and funding
- R01 GM058815/GM/NIGMS NIH HHS/United States
- ER62862/PHS HHS/United States
- GM58815/GM/NIGMS NIH HHS/United States
- HG002318/HG/NHGRI NIH HHS/United States
- R01 HG002318/HG/NHGRI NIH HHS/United States
- CA094816/CA/NCI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous