The Atlas genome assembly system - PubMed (original) (raw)
The Atlas genome assembly system
Paul Havlak et al. Genome Res. 2004 Apr.
Abstract
Atlas is a suite of programs developed for assembly of genomes by a "combined approach" that uses DNA sequence reads from both BACs and whole-genome shotgun (WGS) libraries. The BAC clones afford advantages of localized assembly with reduced computational load, and provide a robust method for dealing with repeated sequences. Inclusion of WGS sequences facilitates use of different clone insert sizes and reduces data production costs. A core function of Atlas software is recruitment of WGS sequences into appropriate BACs based on sequence overlaps. Because construction of consensus sequences is from local assembly of these reads, only small (<0.1%) units of the genome are assembled at a time. Once assembled, each BAC is used to derive a genomic layout. This "sequence-based" growth of the genome map has greater precision than with non-sequence-based methods. Use of BACs allows correction of artifacts due to repeats at each stage of the process. This is aided by ancillary data such as BAC fingerprint, other genomic maps, and syntenic relations with other genomes. Atlas was used to assemble a draft DNA sequence of the rat genome; its major components including overlapper and split-scaffold are also being used in pure WGS projects.
Figures
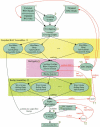
Figure 1
Steps in the Atlas Assembly System. (1) Trim off vector and low-quality portions of reads. (2) Count _k_-mers in WGS reads, saving the overall distribution plus specific counts for _k_-mers with copy number above a threshold. (3) Align BAC and WGS reads sharing rare _k_-mers and save overlap edges for high-quality end-to-end alignments. (4) Enrich each BAC read set with overlapping WGS reads and their mates, assemble using Phrap, scaffold and check for consistent assembly. (5) Arrange BACs into contiguous sets (bactigs) and flag problem BACs for closer quality checking. (6) Assemble bactigs in waves designed to limit the number of BACs that are input to Phrap. (7) Treating each bactig scaffold as a unit, rescaffold to produce superbactigs, detecting problem joins and missed merges between bactigs. (8) Link superbactigs together into ultrabactigs based on remaining (single) mate-pair links, fingerprint contigs, markers and synteny with human and mouse genomes. (9) Format chromosome files with contigs separated by strings of _N_s representing gaps. Quality-control feedback steps include (10) examining coassembly scores of problem BACs and removing foreign trays of reads; (11) resolving superbactig conflicts by modifying bactigs and possibly flagging BACs for closer checking; and (12) resolving ultrabactig and mapping conflicts in collaboration with research groups that generated FPC and marker information.
Figure 2
Read trimming method. A simplified version of the trimming method is shown, with a window size of 4 and a minimum boundary quality of 2. The actual trimming of the rat genome reads used a window size of 50, a minimum boundary quality of 20, and also imposed other requirements on passing windows and regions.
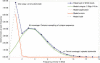
Figure 3
_k_-mer analysis of WGS reads in the RGSP. The frequency distribution of distinct 32-mer oligonucleotides is shown. The observed distribution is shown as a blue line, and the predicted Poisson distribution of unique 32-mers at 4 × shotgun sequencing coverage is shown as the green line. Models for unique 32-mers resulting from sequencing errors, 32-mers in duplicated regions, and the total 32-mer distribution from these models are shown as orange, pink, and dotted lines.
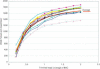
Figure 4
Recruitment of WGS reads into eBACs with increasing BAC skim coverage. Twelve BACs were selected randomly from different chromosomes and used to BAC-Fish WGS reads from a pool representing ∼4 × sequence coverage of the genome. Progressively larger subsets of BAC reads were used to obtain the curves.
Figure 5
False clone overlaps caused by repeated sequences. Each thin line represents a BAC clone, and clones with the same color are true overlapping pairs. False overlaps between clones (lines with different colors) are due to highly repeated sequences as well as duplications of small and large regions. Overlaps from highly repetitious sequences are largely dealt with by the overlapper and binner steps and further dealt with at the overlapping BAC detection step. The second and the third situations are dealt with at the bactig linearization step because conflicts can be detected between BACs. For example, in the second case, A/D and B/D clone pairs should overlap based on the clone layout, but this will not be validated.
Figure 6
The rolling-phrap process limits the scope of reads presented to Phrap in each wave. All reads for an eBAC are added in the first wave including that eBAC. Contigs are recorded in an .ace file, and the corresponding reads are removed from the Phrap input, until a contig contains (almost) no reads from eBACs overlapping the next wave. (1) First wave containing all eBACs overlapping leftmost eBAC (arrow); (2) emit pure leftmost-eBAC contigs (not overlapping and therefore not merged with any other eBAC); (3) second wave containing all eBACs overlapping next leftmost eBAC contributing new sequences; (4) emit contigs solely from first-wave eBAC regions; (5) third wave containing all eBACs overlapping next leftmost eBAC contributing new sequences; (6) emit new contigs solely from first and second-wave eBAC regions; (7) fourth wave; (8) emit remaining contigs.
Similar articles
- Dynamic building of a BAC clone tiling path for the Rat Genome Sequencing Project.
Chen R, Sodergren E, Weinstock GM, Gibbs RA. Chen R, et al. Genome Res. 2004 Apr;14(4):679-84. doi: 10.1101/gr.2171704. Genome Res. 2004. PMID: 15060010 Free PMC article. - LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes.
Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. Frenkel Z, et al. BMC Bioinformatics. 2010 Nov 30;11:584. doi: 10.1186/1471-2105-11-584. BMC Bioinformatics. 2010. PMID: 21118513 Free PMC article. - Bos taurus genome assembly.
Liu Y, Qin X, Song XZ, Jiang H, Shen Y, Durbin KJ, Lien S, Kent MP, Sodeland M, Ren Y, Zhang L, Sodergren E, Havlak P, Worley KC, Weinstock GM, Gibbs RA. Liu Y, et al. BMC Genomics. 2009 Apr 24;10:180. doi: 10.1186/1471-2164-10-180. BMC Genomics. 2009. PMID: 19393050 Free PMC article. - Physical map-assisted whole-genome shotgun sequence assemblies.
Warren RL, Varabei D, Platt D, Huang X, Messina D, Yang SP, Kronstad JW, Krzywinski M, Warren WC, Wallis JW, Hillier LW, Chinwalla AT, Schein JE, Siddiqui AS, Marra MA, Wilson RK, Jones SJ. Warren RL, et al. Genome Res. 2006 Jun;16(6):768-75. doi: 10.1101/gr.5090606. Genome Res. 2006. PMID: 16741162 Free PMC article. - BACFinder: genomic localisation of large insert genomic clones based on restriction fingerprinting.
Crowe ML, Rana D, Fraser F, Bancroft I, Trick M. Crowe ML, et al. Nucleic Acids Res. 2002 Nov 1;30(21):e118. doi: 10.1093/nar/gnf117. Nucleic Acids Res. 2002. PMID: 12409477 Free PMC article.
Cited by
- Current status and future perspectives for sequencing livestock genomes.
Bai Y, Sartor M, Cavalcoli J. Bai Y, et al. J Anim Sci Biotechnol. 2012 Mar 1;3(1):8. doi: 10.1186/2049-1891-3-8. J Anim Sci Biotechnol. 2012. PMID: 22958500 Free PMC article. - Two-step genomic sequence comparison strategy to design Trichoderma strain-specific primers for quantitative PCR.
Zhang Y, Wang X, Pang G, Cai F, Zhang J, Shen Z, Li R, Shen Q. Zhang Y, et al. AMB Express. 2019 Nov 9;9(1):179. doi: 10.1186/s13568-019-0904-4. AMB Express. 2019. PMID: 31707479 Free PMC article. - Improving genome assemblies and annotations for nonhuman primates.
Norgren RB Jr. Norgren RB Jr. ILAR J. 2013;54(2):144-53. doi: 10.1093/ilar/ilt037. ILAR J. 2013. PMID: 24174438 Free PMC article. Review. - Genome-to-phenome research in rats: progress and perspectives.
Zinski AL, Carrion S, Michal JJ, Gartstein MA, Quock RM, Davis JF, Jiang Z. Zinski AL, et al. Int J Biol Sci. 2021 Jan 1;17(1):119-133. doi: 10.7150/ijbs.51628. eCollection 2021. Int J Biol Sci. 2021. PMID: 33390838 Free PMC article. Review. - Quantification of Azospirillum brasilense FP2 Bacteria in Wheat Roots by Strain-Specific Quantitative PCR.
Stets MI, Alqueres SM, Souza EM, Pedrosa Fde O, Schmid M, Hartmann A, Cruz LM. Stets MI, et al. Appl Environ Microbiol. 2015 Oct;81(19):6700-9. doi: 10.1128/AEM.01351-15. Epub 2015 Jul 17. Appl Environ Microbiol. 2015. PMID: 26187960 Free PMC article.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. - PubMed
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. - PubMed
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796-815. - PubMed
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. - PubMed
WEB SITE REFERENCES
- http://www.hgsc.bcm.tmc.edu/BAC-Fisher; BAC-Fisher.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources