S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway - PubMed (original) (raw)
S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway
Mario Pende et al. Mol Cell Biol. 2004 Apr.
Abstract
Activation of 40S ribosomal protein S6 kinases (S6Ks) is mediated by anabolic signals triggered by hormones, growth factors, and nutrients. Stimulation by any of these agents is inhibited by the bacterial macrolide rapamycin, which binds to and inactivates the mammalian target of rapamycin, an S6K kinase. In mammals, two genes encoding homologous S6Ks, S6K1 and S6K2, have been identified. Here we show that mice deficient for S6K1 or S6K2 are born at the expected Mendelian ratio. Compared to wild-type mice, S6K1(-/-) mice are significantly smaller, whereas S6K2(-/-) mice tend to be slightly larger. However, mice lacking both genes showed a sharp reduction in viability due to perinatal lethality. Analysis of S6 phosphorylation in the cytoplasm and nucleoli of cells derived from the distinct S6K genotypes suggests that both kinases are required for full S6 phosphorylation but that S6K2 may be more prevalent in contributing to this response. Despite the impairment of S6 phosphorylation in cells from S6K1(-/-)/S6K2(-/-) mice, cell cycle progression and the translation of 5'-terminal oligopyrimidine mRNAs were still modulated by mitogens in a rapamycin-dependent manner. Thus, the absence of S6K1 and S6K2 profoundly impairs animal viability but does not seem to affect the proliferative responses of these cell types. Unexpectedly, in S6K1(-/-)/S6K2(-/-) cells, S6 phosphorylation persisted at serines 235 and 236, the first two sites phosphorylated in response to mitogens. In these cells, as well as in rapamycin-treated wild-type, S6K1(-/-), and S6K2(-/-) cells, this step was catalyzed by a mitogen-activated protein kinase (MAPK)-dependent kinase, most likely p90rsk. These data reveal a redundancy between the S6K and the MAPK pathways in mediating early S6 phosphorylation in response to mitogens.
Figures
FIG. 1.
Generation of an _S6K2_-null allele. (A) Murine S6K2 gene map and targeting vector. Rectangles represent the coding exons of the S6K2 gene as well as the neomycin resistance (Neo) and thymidine kinase (TK) genes. The exons containing the T loop and APE sequences and the initiation and stop codons are indicated. The four exons deleted following the homologous recombination event are shown in grey. Restriction sites: B, BamHI; Sp, SpeI; E, EcoRI; P, PstI; Sm, SmaI. (B) Genotypes of littermates from S6K2+/− crosses. Tail DNA was digested with BamHI, analyzed by Southern blotting, and hybridized with the probe depicted in panel A. The corresponding BamHI fragments of genomic DNA are 9.8 kb in the wild-type allele and 8.2 kb in the targeted allele. (C and D) Loss of S6K2 protein and kinase activity in S6K2 homozygous mutant cells. (C) Western blot (WB) analysis of fibroblast cell extracts from wild-type or _S6K2_−/− embryos with a MAb against the N-terminal (N-term) region of S6K2. The apparent molecular mass of S6K2 is 68 kDa. (D) MEFs from wild-type or _S6K2_−/− embryos were starved overnight and either stimulated with 10% FCS for 30 min or left untreated. Equal amounts of protein extracts were immunoprecipitated (IP) with the anti-S6K2 antibody, and S6K activity was measured with an immune complex assay (see Materials and Methods).
FIG. 2.
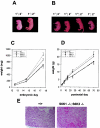
Phenotypes of _S6K1_−/−/_S6K2_−/− mice. (A) Littermates from an _S6K1_−/−/S6K2+/− cross a few minutes after natural delivery. The genotypes of the mice are indicated. The _S6K1_−/−/_S6K2_−/− mouse was born dead and left in the yolk sac. (B) Littermates from an S6K1+/−/_S6K2_−/− cross at embryonic day 19.5 a few minutes after caesarean delivery. The genotypes of the mice are indicated. _S6K1_−/−/_S6K2_−/− mice were more prone to develop transient signs of cyanosis. (C and D) Fetal (C) and postnatal (D) growth curves for mice from homozygous crosses. Values for mice with the same genotype did not differ from those for mice derived from heterozygous crosses. Data represent the average and standard deviation for at least 10 mice per genotype. wt, wild type. (E) Heart histologic findings for wild-type and _S6K1_−/−/_S6K2_−/− mice a few minutes after delivery. The _S6K1_−/−/_S6K2_−/− mouse was born dead. Note hemorrhages in the myocardium.
FIG. 3.
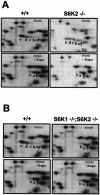
Regulation of S6 phosphorylation. (A) In _S6K2_−/− cells, S6 phosphorylation was reduced yet regulated by mitogens and inhibited by rapamycin (Rapa). 2D PAGE was carried out with 80S ribosomal proteins from wild-type and _S6K2_−/− MEFs. Cells were stimulated with 10% FCS for 1 h, with or without pretreatment with 20 nM rapamycin. (B) S6 phosphorylation in _S6K1_−/−/_S6K2_−/− cells was reduced to the same extent as in rapamycin-treated cells. 2D PAGE was carried out with 80S ribosomal proteins from wild-type and _S6K1_−/−/_S6K2_−/− MEFs. Cells were treated as described for panel A. See the text for an explanation of a to e on the panels.
FIG. 4.
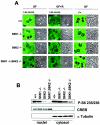
Intracellular localization of S6K activity. (A) Immunostaining of wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes with anti-phosphorylated S6 (Ser235/236) and anti-L7a antibodies (left panels). The right panels represent phase-contrast images of the same fields. Cells were growth factor deprived for 1 day and stimulated with 4 nM EGF (25 ng/ml) and 1 μM insulin for 1 h (GF), with or without pretreatment with 20 nM rapamycin (R). Arrowheads indicate nucleoli. (B) Immunoblot analysis of nuclear and cytosolic extracts from wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes with anti-phosphorylated S6 (Ser235/236), anti-α-tubulin, and anti-CREB antibodies. The latter two were used as cytosolic and nuclear markers, respectively.
FIG. 5.
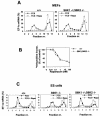
Regulation of 5′TOP mRNA translation and cell cycle progression is preserved in _S6K1_−/−/_S6K2_−/− MEFs. (A and C) 5′TOP mRNA translation is modulated by serum and rapamycin (Rapa), although S6 phosphorylation is resistant to rapamycin treatment. Cytoplasmic extracts were prepared from wild-type (+/+), _S6K1_−/−, and _S6K1_−/−/_S6K2_−/− MEFs (A) or ES cells (C) that had been stimulated with 10% serum (FCS) in the presence (empty circles) or absence (filled circles) of 20 nM rapamycin after serum deprivation for 48 h. Extracts were centrifuged on a sucrose gradient prior to fractionation as previously described (22). RNA from the different fractions was analyzed by Northern blotting with a probe specific for EF1α (EF-1a) mRNA. The relative amount of mRNA present in each fraction is expressed as a percentage. Fractions 1 to 7 contain polysomes, whereas fractions 8 to 14 are enriched in monosomes, ribosomal subunits, and mRNPs. nr., number. (B) Inhibitory effect of rapamycin on the proliferation of S6K-null cells. Wild-type MEFs (filled circles) and _S6K1_−/−/_S6K2_−/− MEFs (empty circles) were seeded in a 24-well plate at a density of 2 × 104 cells per well. Cells then were treated with 0.5% serum for 48 h and stimulated for 20 h with 10% serum in the absence or presence of rapamycin concentrations ranging from 0 to 100 nM. Cells were labeled with [3H]thymidine (1 μCi/ml) throughout the entire stimulation period Radioactivity incorporated (incorp.) into the DNA was measured, and the average values for triplicate samples were determined. Data are expressed as the percent inhibition of [3H]thymidine incorporation by rapamycin-treated cells compared to untreated cells.
FIG. 6.
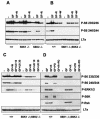
Evidence for a rapamycin-insensitive S6K. Western blot analysis of protein extracts from wild-type, _S6K1_−/−, _S6K2_−/−, and _S6K1_−/−/_S6K2_−/− hepatocytes was carried out with anti-phosphorylated S6 (Ser235/236 and Ser240/244), anti-phosphorylated ERK1/ERK2, anti-phosphorylated S6K (Thr389), anti-phosphorylated Rsk (Thr359 and Ser363), and anti-L7a antibodies, the latter being used as a loading control. Cells were stimulated with 4 nM EGF (25 ng/ml) and 1 μM insulin for 30 min (GF), with or without pretreatment with 20 nM rapamycin (R), 1 μM wortmannin (W), 10 or 30 μM U0126 (U0 10 or U0 30, respectively), and 3 μM PD184352 (PD). Extracts in panels A and C were derived from cells deprived of GFs for 1 day, whereas those in panels B and D were derived from cells deprived of GFs for 2 days. In panel D, cells were also deprived of nutrients (all of the amino acids and glucose) for 3 h.
Similar articles
- mGluR-dependent long-term depression is associated with increased phosphorylation of S6 and synthesis of elongation factor 1A but remains expressed in S6K-deficient mice.
Antion MD, Hou L, Wong H, Hoeffer CA, Klann E. Antion MD, et al. Mol Cell Biol. 2008 May;28(9):2996-3007. doi: 10.1128/MCB.00201-08. Epub 2008 Mar 3. Mol Cell Biol. 2008. PMID: 18316404 Free PMC article. - Mutational analysis of ribosomal S6 kinase 2 shows differential regulation of its kinase activity from that of ribosomal S6 kinase 1.
Phin S, Kupferwasser D, Lam J, Lee-Fruman KK. Phin S, et al. Biochem J. 2003 Jul 15;373(Pt 2):583-91. doi: 10.1042/BJ20021794. Biochem J. 2003. PMID: 12713446 Free PMC article. - Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation.
Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O. Stolovich M, et al. Mol Cell Biol. 2002 Dec;22(23):8101-13. doi: 10.1128/MCB.22.23.8101-8113.2002. Mol Cell Biol. 2002. PMID: 12417714 Free PMC article. - Ribosomal protein S6 kinase from TOP mRNAs to cell size.
Meyuhas O, Dreazen A. Meyuhas O, et al. Prog Mol Biol Transl Sci. 2009;90:109-53. doi: 10.1016/S1877-1173(09)90003-5. Epub 2009 Oct 27. Prog Mol Biol Transl Sci. 2009. PMID: 20374740 Review. - Role of S6 phosphorylation and S6 kinase in cell growth.
Volarević S, Thomas G. Volarević S, et al. Prog Nucleic Acid Res Mol Biol. 2001;65:101-27. doi: 10.1016/s0079-6603(00)65003-1. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11008486 Review.
Cited by
- Unique and Redundant Functions of p70 Ribosomal S6 Kinase Isoforms Regulate Mesenchymal Cell Proliferation and Migration in Pulmonary Fibrosis.
Madala SK, Sontake V, Edukulla R, Davidson CR, Schmidt S, Hardie WD. Madala SK, et al. Am J Respir Cell Mol Biol. 2016 Dec;55(6):792-803. doi: 10.1165/rcmb.2016-0090OC. Am J Respir Cell Mol Biol. 2016. PMID: 27438654 Free PMC article. - Muscle fiber type-dependent differences in the regulation of protein synthesis.
Goodman CA, Kotecki JA, Jacobs BL, Hornberger TA. Goodman CA, et al. PLoS One. 2012;7(5):e37890. doi: 10.1371/journal.pone.0037890. Epub 2012 May 22. PLoS One. 2012. PMID: 22629468 Free PMC article. - RNA interference-mediated repression of S6 kinase 1 impairs root nodule development in soybean.
Um JH, Kim S, Kim YK, Song SB, Lee SH, Verma DP, Cheon CI. Um JH, et al. Mol Cells. 2013 Mar;35(3):243-8. doi: 10.1007/s10059-013-2315-8. Epub 2013 Mar 8. Mol Cells. 2013. PMID: 23475423 Free PMC article. - Impact of periconceptional and preimplantation undernutrition on factors regulating myogenesis and protein synthesis in muscle of singleton and twin fetal sheep.
Lie S, Morrison JL, Williams-Wyss O, Suter CM, Humphreys DT, Ozanne SE, Zhang S, MacLaughlin SM, Kleemann DO, Walker SK, Roberts CT, McMillen IC. Lie S, et al. Physiol Rep. 2015 Aug;3(8):e12495. doi: 10.14814/phy2.12495. Physiol Rep. 2015. PMID: 26265755 Free PMC article. - Translational control of myelin basic protein expression by ERK2 MAP kinase regulates timely remyelination in the adult brain.
Michel K, Zhao T, Karl M, Lewis K, Fyffe-Maricich SL. Michel K, et al. J Neurosci. 2015 May 20;35(20):7850-65. doi: 10.1523/JNEUROSCI.4380-14.2015. J Neurosci. 2015. PMID: 25995471 Free PMC article.
References
- Abraham, R. T. 2002. Identification of TOR signaling complexes: more TORC for the cell growth engine. Cell 111:9-12. - PubMed
- Arber, S., J. J. Hunter, J. Ross, Jr., M. Hongo, G. Sansig, J. Borg, J. C. Perriard, K. R. Chien, and P. Caroni. 1997. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell 88:393-403. - PubMed
- Bandi, H. R., S. Ferrari, J. Krieg, H. E. Meyer, and G. Thomas. 1993. Identification of 40 S ribosomal protein S6 phosphorylation sites in Swiss mouse 3T3 fibroblasts stimulated with serum. J. Biol. Chem. 268:4530-4533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases