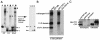Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance - PubMed (original) (raw)
Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance
Vinod Pant et al. Mol Cell Biol. 2004 Apr.
Abstract
The differentially methylated imprinting control region (ICR) region upstream of the H19 gene regulates allelic Igf2 expression by means of a methylation-sensitive chromatin insulator function. We have previously shown that maternal inheritance of mutated (three of the four) target sites for the 11-zinc finger protein CTCF leads to loss of Igf2 imprinting. Here we show that a mutation in only CTCF site 4 also leads to robust activation of the maternal Igf2 allele despite a noticeably weaker interaction in vitro of site 4 DNA with CTCF compared to other ICR sites, sites 1 and 3. Moreover, maternally inherited sites 1 to 3 become de novo methylated in complex patterns in subpopulations of liver and heart cells with a mutated site 4, suggesting that the methylation privilege status of the maternal H19 ICR allele requires an interdependence between all four CTCF sites. In support of this conclusion, we show that CTCF molecules bind to each other both in vivo and in vitro, and we demonstrate strong interaction between two CTCF-DNA complexes, preassembled in vitro with sites 3 and 4. We propose that the CTCF sites may cooperate to jointly maintain both methylation-free status and insulator properties of the maternal H19 ICR allele. Considering many other CTCF targets, we propose that site-specific interactions between various DNA-bound CTCF molecules may provide general focal points in the organization of looped chromatin domains involved in gene regulation.
Figures
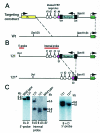
FIG. 1.
Schematic illustration of the knock-in strategy. (A) Recombination map of the targeting construct and endogenous H19 ICR. CTCF target site 4 (cerise box) was targeted in the knock-in procedure. Wt, wild-type. (B) Deletion of the neomycin gene by breeding mice of strain 121 with mice harboring a β-actin promoter-driven Cre recombinase gene to generate the 121* substrain. (C) The recombination event in clone 121 was assessed by using a probe that is positioned 5′ of the sequence covered in the targeting construct. The properly recombined insert generated a larger fragment due to the extra sequence information provided by the neomycin gene. The mutated targeting allele replaced the endogenous sequence in clone 121 as confirmed by EcoRV (EV) digestion and Southern blot analysis. The mutated allele of the 121* strain is indistinguishable from the wild-type allele when digested with BamHI (B) and DraI (D). For additional information, see the text.
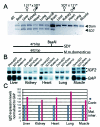
FIG. 2.
Expression analyses of Igf2. (A) RT-PCR analyses of Igf2 expression pattern in various organs dissected from neonatal offspring after reciprocal crosses between 121 and SD7 mice. A polymorphic BsaAI site in exon 4 of Igf2, present only in SD7 mice, was used to discriminate between the alleles. The animal crosses are presented in the order of female to male. The −RT lane contained RNA and primers but no reverse transcriptase for a control. (B) Northern blot analysis of Igf2 expression in neonatal mice harboring the mutated allele on the maternal chromosome. The blot was stripped and reprobed with the glyceraldehyde-3-phosphate dehydrogenase (GAP) probe to confirm RNA loading. (C) Graph showing Igf2 expression after the levels were normalized to GAP expression. The level of wild-type Igf2 expression was set at 100%. Contr., contributed; Mat. inher., maternally inherited.
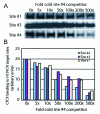
FIG. 3.
Binding efficiencies of the H19 ICR CTCF target sites. (A) CTCF target sites 1, 2, and 4 were labeled with 32P and competed with cold site 4 in band shift analyses. (B) The graph depicts the mean values for two independent experiments describing the degree of competition on individual sites in relation to fold increases in the amount of the cold competitor.
FIG. 4.
CpG methylation analyses. (A) Southern blot analysis of genomic DNA isolated from various neonatal organs of reciprocal crosses between 121 and SD7 mice. The various tissues were digested with HhaI (+) or not digested with HhaI (−). The positions of molecular size markers (in kilobases) are indicated to the right of the blots. The parental alleles were discriminated by exploiting a polymorphic BstXI site, which is shown in the map below the blots. (B) Bisulfite sequencing analyses of the maternally transmitted mutant H19 ICR allele covering all four CTCF target sites. Region A contained sites 1 and 2, while region B contained sites 3 and 4. The wild-type and mutant alleles were discriminated by known single-base polymorphism in region A and by the introduced point mutation in region B. Filled circles represent methylated CpGs, and open circles represent unmethylated CpGs. (C) Graphical representation of the percentage of individual CpG methylation in H19 ICR.
FIG. 5.
CTCF interacts with itself in vivo and in vitro, while only heterologous CTCF-DNA complexes interact in vitro. (A) DNA fragments containing CTCF target sites 3 and 4 were labeled with 32P and subjected to CTCF band shift analysis either separately or in combination. Shifted band fragments (bands 1, 2, and 3) were eluted from the gel (left gel), treated with proteinase K, and resolved on a 5% acrylamide gel (right gel). F, free; B, bound. (B) The C terminus of CTCF binds to the 11 ZF domain of CTCF. The latter was initially noticed in the preliminary pull-down assay with various portions of CTCF (data not shown). Next, the IVT radiolabeled full-length CTCF, the 11 ZF domain, and the CTCF-C domain were mixed together at a ratio of approximately 1:1:1 as shown in the Input lane and subjected to a GST pull-down assay with the three bacterially expressed GST fusion proteins, GST-ABL, GST-CTCF-C, and GST-CTCF-N, immobilized on glutathione beads. (C) CTCF interacts with itself in vivo. COS 7 cells were transfected with _myc_-tagged CTCF, and the _myc_-tagged (9E10) antibody was used for coimmunoprecipitation. Immunoprecipitates (Immunoprec.) and control lysates were resolved on SDS-10% polyacrylamide gels and probed with the anti-CTCF antibody. The positions of the endogenous CTCF (16) and _myc_-tagged exogenous CTCF (15) are indicated to the left of the blot. No signal was observed in the controls obtained with the preimmune serum and anti-α-tubulin antibody.
Similar articles
- CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2.
Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. Kurukuti S, et al. Proc Natl Acad Sci U S A. 2006 Jul 11;103(28):10684-9. doi: 10.1073/pnas.0600326103. Epub 2006 Jun 30. Proc Natl Acad Sci U S A. 2006. PMID: 16815976 Free PMC article. - Complete biallelic insulation at the H19/Igf2 imprinting control region position results in fetal growth retardation and perinatal lethality.
Lee DH, Singh P, Tsark WM, Szabó PE. Lee DH, et al. PLoS One. 2010 Sep 8;5(9):e12630. doi: 10.1371/journal.pone.0012630. PLoS One. 2010. PMID: 20838620 Free PMC article. - CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia.
Lee DH, Singh P, Tsai SY, Oates N, Spalla A, Spalla C, Brown L, Rivas G, Larson G, Rauch TA, Pfeifer GP, Szabó PE. Lee DH, et al. PLoS Genet. 2010 Nov 24;6(11):e1001224. doi: 10.1371/journal.pgen.1001224. PLoS Genet. 2010. PMID: 21124827 Free PMC article. - Genomic imprinting: CTCF protects the boundaries.
Lewis A, Murrell A. Lewis A, et al. Curr Biol. 2004 Apr 6;14(7):R284-6. doi: 10.1016/j.cub.2004.03.026. Curr Biol. 2004. PMID: 15062124 Review. - CTCF function is modulated by neighboring DNA binding factors.
Weth O, Renkawitz R. Weth O, et al. Biochem Cell Biol. 2011 Oct;89(5):459-68. doi: 10.1139/o11-033. Epub 2011 Sep 7. Biochem Cell Biol. 2011. PMID: 21895576 Review.
Cited by
- CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention.
Pugacheva EM, Kubo N, Loukinov D, Tajmul M, Kang S, Kovalchuk AL, Strunnikov AV, Zentner GE, Ren B, Lobanenkov VV. Pugacheva EM, et al. Proc Natl Acad Sci U S A. 2020 Jan 28;117(4):2020-2031. doi: 10.1073/pnas.1911708117. Epub 2020 Jan 14. Proc Natl Acad Sci U S A. 2020. PMID: 31937660 Free PMC article. - Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus.
Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, Pasini D, Miró X, Donohoe ME, Lee JT, Helin K, Soloway PD. Lindroth AM, et al. PLoS Genet. 2008 Aug 1;4(8):e1000145. doi: 10.1371/journal.pgen.1000145. PLoS Genet. 2008. PMID: 18670629 Free PMC article. - Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions.
Bushey AM, Ramos E, Corces VG. Bushey AM, et al. Genes Dev. 2009 Jun 1;23(11):1338-50. doi: 10.1101/gad.1798209. Epub 2009 May 14. Genes Dev. 2009. PMID: 19443682 Free PMC article. - Assessing Self-interaction of Mammalian Nuclear Proteins by Co-immunoprecipitation.
Cattoglio C, Pustova I, Darzacq X, Tjian R, Hansen AS. Cattoglio C, et al. Bio Protoc. 2020 Feb 20;10(4):e3526. doi: 10.21769/BioProtoc.3526. eCollection 2020 Feb 20. Bio Protoc. 2020. PMID: 33654750 Free PMC article. - Epigenomic regulation of heart failure: integrating histone marks, long noncoding RNAs, and chromatin architecture.
McKinsey TA, Vondriska TM, Wang Y. McKinsey TA, et al. F1000Res. 2018 Oct 29;7:F1000 Faculty Rev-1713. doi: 10.12688/f1000research.15797.1. eCollection 2018. F1000Res. 2018. PMID: 30416708 Free PMC article. Review.
References
- Bartolomei, M. S., and S. M. Tilghman. 1997. Genomic imprinting in mammals. Annu. Rev. Genet. 31:493-525. - PubMed
- Bell, A., A. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
- Chernukhin, I., S. Shamsuddin, A. Robinson, A. Carne, A. Paul, A. El-Kady, V. Lobanenkov, and E. Klenova. 2000. Physical and functional interaction between two pluripotent proteins, the Y-box DNA/RNA-binding factor, YB-1, and the multivalent zinc finger factor, CTCF. J. Biol. Chem. 275:29915-29921. - PubMed
- Davis, T., G. Yang, J. McCarrey, and M. Bartolomei. 2000. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 9:2885-2894. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous