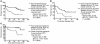Gene expression profiling predicts clinical outcome of prostate cancer - PubMed (original) (raw)
Gene expression profiling predicts clinical outcome of prostate cancer
Gennadi V Glinsky et al. J Clin Invest. 2004 Mar.
Abstract
One of the major problems in management of prostate cancer is the lack of reliable genetic markers predicting the clinical course of the disease. We analyzed expression profiles of 12,625 transcripts in prostate tumors from patients with distinct clinical outcomes after therapy as well as metastatic human prostate cancer xenografts in nude mice. We identified small clusters of genes discriminating recurrent versus nonrecurrent disease with 90% and 75% accuracy in two independent cohorts of patients. We examined one group of samples (21 tumors) to discover the recurrence predictor genes and then validated the predictive power of these genes in a different set (79 tumors). Kaplan-Meier analysis demonstrated that recurrence predictor signatures are highly informative (P < 0.0001) in stratification of patients into subgroups with distinct relapse-free survival after therapy. A gene expression-based recurrence predictor algorithm was informative in predicting the outcome in patients with early-stage disease, with either high or low preoperative prostate-specific antigen levels and provided additional value to the outcome prediction based on Gleason sum or multiparameter nomogram. Overall, 88% of patients with recurrence of prostate cancer within 1 year after therapy were correctly classified into the poor-prognosis group. The identified algorithm provides additional predictive value over conventional markers of outcome and appears suitable for stratification of prostate cancer patients at the time of diagnosis into subgroups with distinct survival probability after therapy.
Figures
Figure 1
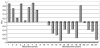
PAIs defined by the expression profile of the prostate cancer recurrence predictor signature 1 for 21 prostate carcinoma samples constituting a signature discovery (training) data set. Prostate tumor samples were taken from the patients at the time of surgery and subjected to a microarray gene expression analysis as described in Methods. Note that all samples derived from tumors of patients who subsequently manifested a biochemical relapse of disease have positive PAI values, whereas 12 of 13 samples obtained from patients who remained disease-free have negative PAI values. See text for details.
Figure 2
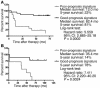
Kaplan-Meier analysis of the probability that patients would remain disease-free among 21 prostate cancer patients constituting a signature discovery group according to whether they had good-prognosis or poor-prognosis signatures defined by the recurrence predictor signature 1 (A), recurrence predictor signature 2 (B), recurrence predictor signature 3 (C), and the recurrence predictor algorithm, which takes into account calls from all three signatures (D). The cut-off values for each marker were identified through the detailed analysis of behavior of log-rank test P values across the range of the measurements for each marker. We selected the prognosis discrimination cut-off value for each signature based on highest level of statistical significance in patients’ stratification into poor- and good-prognosis groups as determined by the log-rank test (lowest P value and highest hazard ratio; see Supplemental Table 6S). CI, confidence interval.
Figure 3
Kaplan-Meier analysis of the probability that patients would remain disease-free among 79 prostate cancer patients constituting a signature validation group for all patients (A), patients with high (B) or low (C) preoperative PSA levels in blood according to whether they had good-prognosis or poor-prognosis signatures defined by the recurrence predictor algorithm, or whether they had high or low preoperative PSA level in the blood (D). Preoperative PSA level of 7.8 ng/ml was used as a cut-off discrimination level for patients’ stratification into poor- and good-prognosis subgroups. The cut-off values for each marker were identified through the detailed analysis of behavior of log-rank test P values across the range of the measurements for each marker. We selected the prognosis discrimination cut-off value for each marker based on highest level of statistical significance in patients’ stratification into poor- and good-prognosis groups as determined by the log-rank test (lowest P value and highest hazard ratio).
Figure 4
Kaplan-Meier analysis of the probability that patients would remain disease-free among prostate cancer patients with Gleason sum 6 and 7 tumors (A) and patients with Gleason sum 8 and 9 tumors (B) according to whether they had good-prognosis or poor-prognosis signatures defined by the recurrence predictor algorithm or whether they had Gleason sum 8 and 9 or Gleason sum 6 and 7 prostate tumors (C).
Figure 5
Kaplan-Meier analysis of the probability that patients would remain disease-free among 79 prostate cancer patients constituting a signature validation group for all patients (A), patients with poor prognosis (B), or good prognosis (C), defined by the Kattan nomogram according to whether they had a good-prognosis or poor-prognosis signatures defined by the recurrence predictor algorithm (B and C) or whether they had poor or good prognosis defined by the Kattan nomogram (A).
Figure 6
Kaplan-Meier analysis of the probability that patients would remain disease-free among prostate cancer patients with stage 1C tumors (A) and patients with stage 2A tumors (B) according to whether they had a good-prognosis or poor-prognosis signatures defined by the recurrence predictor algorithm.
Comment in
- Predicting the clinical course of prostate cancer.
McKiernan J, Benson MC. McKiernan J, et al. J Clin Invest. 2004 Mar;113(6):806-8. doi: 10.1172/JCI21310. J Clin Invest. 2004. PMID: 15067311 Free PMC article.
Similar articles
- A genomic classifier improves prediction of metastatic disease within 5 years after surgery in node-negative high-risk prostate cancer patients managed by radical prostatectomy without adjuvant therapy.
Klein EA, Yousefi K, Haddad Z, Choeurng V, Buerki C, Stephenson AJ, Li J, Kattan MW, Magi-Galluzzi C, Davicioni E. Klein EA, et al. Eur Urol. 2015 Apr;67(4):778-86. doi: 10.1016/j.eururo.2014.10.036. Epub 2014 Nov 12. Eur Urol. 2015. PMID: 25466945 - Integration of gene expression profiling and clinical variables to predict prostate carcinoma recurrence after radical prostatectomy.
Stephenson AJ, Smith A, Kattan MW, Satagopan J, Reuter VE, Scardino PT, Gerald WL. Stephenson AJ, et al. Cancer. 2005 Jul 15;104(2):290-8. doi: 10.1002/cncr.21157. Cancer. 2005. PMID: 15948174 Free PMC article. - Clinical utility of microarray-derived genetic signatures in predicting outcomes in prostate cancer.
Reddy GK, Balk SP. Reddy GK, et al. Clin Genitourin Cancer. 2006 Dec;5(3):187-9. doi: 10.3816/CGC.2006.n.035. Clin Genitourin Cancer. 2006. PMID: 17239271 Review. - Molecular markers to identify patients at risk for recurrence after primary treatment for prostate cancer.
Kumar-Sinha C, Chinnaiyan AM. Kumar-Sinha C, et al. Urology. 2003 Dec 29;62 Suppl 1:19-35. doi: 10.1016/j.urology.2003.10.007. Urology. 2003. PMID: 14747039 Review.
Cited by
- ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients.
Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, Kim J, Nguyen M, Zhang X, Godinho FJ, Bronson RT, Mucci LA, Loda M, Yuan GC, Orkin SH, Li Z. Baena E, et al. Genes Dev. 2013 Mar 15;27(6):683-98. doi: 10.1101/gad.211011.112. Genes Dev. 2013. PMID: 23512661 Free PMC article. - Clinical significance of EPHX2 deregulation in prostate cancer.
Liu MS, Zhao H, Xu CX, Xie PB, Wang W, Yang YY, Lee WH, Jin Y, Zhou HQ. Liu MS, et al. Asian J Androl. 2021 Jan-Feb;23(1):109-115. doi: 10.4103/aja.aja_34_20. Asian J Androl. 2021. PMID: 32687069 Free PMC article. - The small GTPase ARF3 controls invasion modality and metastasis by regulating N-cadherin levels.
Sandilands E, Freckmann EC, Cumming EM, Román-Fernández A, McGarry L, Anand J, Galbraith L, Mason S, Patel R, Nixon C, Cartwright J, Leung HY, Blyth K, Bryant DM. Sandilands E, et al. J Cell Biol. 2023 Apr 3;222(4):e202206115. doi: 10.1083/jcb.202206115. Epub 2023 Feb 28. J Cell Biol. 2023. PMID: 36880595 Free PMC article. - URG11 Regulates Prostate Cancer Cell Proliferation, Migration, and Invasion.
Pan B, Ye Y, Liu H, Zhen J, Zhou H, Li Y, Qu L, Wu Y, Zeng C, Zhong W. Pan B, et al. Biomed Res Int. 2018 May 31;2018:4060728. doi: 10.1155/2018/4060728. eCollection 2018. Biomed Res Int. 2018. PMID: 29955600 Free PMC article. - Evaluation of protein biomarkers of prostate cancer aggressiveness.
Rizzardi AE, Rosener NK, Koopmeiners JS, Isaksson Vogel R, Metzger GJ, Forster CL, Marston LO, Tiffany JR, McCarthy JB, Turley EA, Warlick CA, Henriksen JC, Schmechel SC. Rizzardi AE, et al. BMC Cancer. 2014 Apr 5;14:244. doi: 10.1186/1471-2407-14-244. BMC Cancer. 2014. PMID: 24708576 Free PMC article.
References
- Holmberg L, et al. A randomized trial comparing radical prostatectomy with watchful waiting in early prostate cancer. N. Engl. J. Med. 2002;347:781–789. - PubMed
- Thomas, G.V., and Loda, M. 2002. Molecular staging of prostate cancer. In Prostate cancer principles & practice. P.W. Kantoff, P.R. Carroll, and A.V. D’Amico, editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 287–303.
- DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. - PubMed
- Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardinom PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J. Natl. Cancer Inst. 1998;90:766–771. - PubMed
- D’Amico AV, et al. Pretreatment nomogram for prostate-specific antigen recurrence after radical prostatectomy or external-beam radiation therapy for clinically localised prostate cancer. J. Clin. Oncol. 1999;17:168–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA084999/CA/NCI NIH HHS/United States
- U01 CA-84999/CA/NCI NIH HHS/United States
- 1 R43 CA-89779/CA/NCI NIH HHS/United States
- R01 CA089827/CA/NCI NIH HHS/United States
- 5R01 CA-89827/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical