Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota - PubMed (original) (raw)
Comparative Study
. 2004 Mar 30;101(13):4596-601.
doi: 10.1073/pnas.0400706101. Epub 2004 Mar 19.
Affiliations
- PMID: 15070763
- PMCID: PMC384792
- DOI: 10.1073/pnas.0400706101
Comparative Study
Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota
John F Rawls et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Animals have developed the means for supporting complex and dynamic consortia of microorganisms during their life cycle. A transcendent view of vertebrate biology therefore requires an understanding of the contributions of these indigenous microbial communities to host development and adult physiology. These contributions are most obvious in the gut, where studies of gnotobiotic mice have disclosed that the microbiota affects a wide range of biological processes, including nutrient processing and absorption, development of the mucosal immune system, angiogenesis, and epithelial renewal. The zebrafish (Danio rerio) provides an opportunity to investigate the molecular mechanisms underlying these interactions through genetic and chemical screens that take advantage of its transparency during larval and juvenile stages. Therefore, we developed methods for producing and rearing germ-free zebrafish through late juvenile stages. DNA microarray comparisons of gene expression in the digestive tracts of 6 days post fertilization germ-free, conventionalized, and conventionally raised zebrafish revealed 212 genes regulated by the microbiota, and 59 responses that are conserved in the mouse intestine, including those involved in stimulation of epithelial proliferation, promotion of nutrient metabolism, and innate immune responses. The microbial ecology of the digestive tracts of conventionally raised and conventionalized zebrafish was characterized by sequencing libraries of bacterial 16S rDNA amplicons. Colonization of germ-free zebrafish with individual members of its microbiota revealed the bacterial species specificity of selected host responses. Together, these studies establish gnotobiotic zebrafish as a useful model for dissecting the molecular foundations of host-microbial interactions in the vertebrate digestive tract.
Figures
Fig. 1.
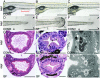
Morphologic studies of CONR, CONV, and GF zebrafish. (A-C) Whole-mount preparations of 6-dpf zebrafish. Rostral is to the left, dorsal is to the top. A shows the position of the swim bladder (SB) and the boundary of intestinal segment 2 (red bracket). Segments 1 and 3 lie rostral and caudal to segment 2, respectively. (D-F) Whole mounts of the caudal regions of 9-dpf CONR, GF, and CONV (conventionalized at 3 dpf) animals, showing onset of epidermal degeneration phenotype in GF fish. This phenotype is manifested by loss of transparency and integrity of the epidermis in fin folds (the edges of these fin folds are highlighted with open arrowheads in E). CONR and CONV fin folds remain transparent (edges indicated by filled black arrowheads in D and F). (G, H, J, and K) Hematoxylin- and eosin-stained transverse sections showing intestinal segment 1 (G and J) and segment 2 (H and K) in 6-dpf CONV and GF zebrafish. There are no detectable epithelial abnormalities in intestinal segment 1, whether judged by light microscopy (G and J) or by transmission EM (data not shown). In contrast, enterocytes in segment 2 contain prominent supranuclear vacuoles filled with eosinophilic material in CONV (and CONR) fish (e.g., black arrowheads in H). These vacuoles appear clear in GF animals (e.g., open arrowheads in K). Pigmented melanocytes (m) lie adjacent to the intestine in H and K. (I and L) EM study of 6-dpf intestines, showing electron-dense material in the supranuclear vacuoles (v) of segment 2 CONV enterocytes, and electron-lucent material in GF enterocytes. The filled black arrowhead in I points to a bacterium in the intestinal lumen. (Bars: 500 μm in A-F; 100 μmin G and J;20 μmin H and K; 5 μmin I and L.)
Fig. 2.
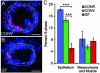
The microbiota stimulates intestinal epithelial proliferation. (A and B) Sections prepared from the intestines of 6-dpf CONV and GF zebrafish after a 24-h exposure to bromodeoxyuridine in their environmental water. Sections were stained with antibodies to bromodeoxyuridine (magenta) and the nuclear stain bisbenzimide (blue). The mesenchyme and muscle surrounding the intestinal epithelium are outlined in white. (C) Quantitation of S-phase cells in the intestinal epithelium and mesenchyme. The percentage of cells in S phase in GF intestinal epithelium is significantly lower than in CONR or CONV animals (P < 0.0001, indicated by brackets with three asterisks). Data are expressed as the mean of two independent experiments ± SEM (n = 19-31 sections scored per animal; ≥6 animals per experiment). (Bars: 25 μm in A and B.)
Fig. 3.
Real-time quantitative RT-PCR studies of the microbial species specificity of selected evolutionarily conserved zebrafish responses to the digestive tract microbiota. Expression levels of serum amyloid A1 (Saa1; A), complement component 3 (C3; B), fasting-induced adipose factor (Fiaf; C), and solute carrier family 31 member 1 (Slc31a1; D) in digestive tracts from 6-dpf conventionalized (CONV), _A. hydrophila_-monoassociated (A.h.), and _P. aeruginosa_-monoassociated (P.a.) larvae are shown relative to 6-dpf GF larval digestive tracts. Assays were performed in triplicate (n ≥ 4 assays per gene). Data were normalized to 18S ribosomal RNA and results are expressed as mean log2 values ± SEM.
References
- McFall-Ngai, M. J. (2002) Dev. Biol. 242, 1-14. - PubMed
- Savage, D. C. (1977) Annu. Rev. Microbiol. 31, 107-133. - PubMed
- Salzman, N. H., de Jong, H., Paterson, Y., Harmsen, H. J., Welling, G. W. & Bos, N. A. (2002) Microbiology 148, 3651-3660. - PubMed
Publication types
MeSH terms
Grants and funding
- DK62675/DK/NIDDK NIH HHS/United States
- R01 DK030292/DK/NIDDK NIH HHS/United States
- DK30292/DK/NIDDK NIH HHS/United States
- F32 DK062675/DK/NIDDK NIH HHS/United States
- R37 DK030292/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases