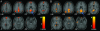Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI - PubMed (original) (raw)
Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI
Michael D Greicius et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Recent functional imaging studies have revealed coactivation in a distributed network of cortical regions that characterizes the resting state, or default mode, of the human brain. Among the brain regions implicated in this network, several, including the posterior cingulate cortex and inferior parietal lobes, have also shown decreased metabolism early in the course of Alzheimer's disease (AD). We reasoned that default-mode network activity might therefore be abnormal in AD. To test this hypothesis, we used independent component analysis to isolate the network in a group of 13 subjects with mild AD and in a group of 13 age-matched elderly controls as they performed a simple sensory-motor processing task. Three important findings are reported. Prominent coactivation of the hippocampus, detected in all groups, suggests that the default-mode network is closely involved with episodic memory processing. The AD group showed decreased resting-state activity in the posterior cingulate and hippocampus, suggesting that disrupted connectivity between these two regions accounts for the posterior cingulate hypometabolism commonly detected in positron emission tomography studies of early AD. Finally, a goodness-of-fit analysis applied at the individual subject level suggests that activity in the default-mode network may ultimately prove a sensitive and specific biomarker for incipient AD.
Figures
Fig. 1.
Validation of the ICA approach (Stanford University data). Axial images showing the default-mode network as detected with ROI-based (A) and ICA-based (B) approaches in a group of healthy young adults scanned on a 3-T magnet at Stanford University. The blue arrows indicate the PCC. The left side of the image corresponds to the left side of the brain. The numbers beneath each image refer to the z coordinate in Talairach space. T score bars are shown at right. Functional images were overlaid on the group-averaged structural image. Joint height and extent thresholds of P < 0.001 were used to determine significant clusters.
Fig. 2.
Hippocampal coactivation in the default-mode network (Washington University data). Axial (A) and coronal (B) images showing the default-mode network for 14 healthy young subjects scanned on a 1.5-T magnet at Washington University. The green arrows highlight the prominent bilateral coactivation in the hippocampus and underlying entorhinal cortex. The numbers beneath each coronal image refer to the y coordinate in Talairach space. Joint height and extent thresholds of P < 0.0001 were used to determine significant clusters. Other details are as in Fig. 1.
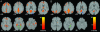
Fig. 3.
Default-mode network in healthy elderly and AD subjects (Washington University data). Axial images showing the default-mode network for the healthy elderly (A) and AD (B) groups. The blue arrows indicate the PCC. The hippocampus and underlying entorhinal cortex (green arrows) were detected bilaterally in healthy elderly subjects (A) but only in the right hemisphere in the AD group (B). Joint height and extent thresholds of P < 0.0001 were used to determine significant clusters. Other details are as in Fig. 1.
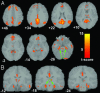
Fig. 4.
Increased default-mode network activity in healthy elderly. (A) Axial images showing the results of a two-sample t test contrasting the default-mode network in the healthy elderly group vs. the AD group. The blue arrow indicates the PCC. The magenta arrows indicate the inferior parietal lobes. (B) Coronal images showing a 112-voxel cluster in the left hippocampus (green arrows) that survived the height but not the extent threshold. Joint height and extent thresholds of P < 0.05 were used to determine significant clusters. Other details are as in Fig. 1.
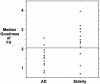
Fig. 5.
Individual scores for goodness of fit to standard default-mode network. A scattergram shows the median goodness of fit for each subject in the AD and healthy elderly groups using the Stanford University ICA-derived default-mode template. The group means were significantly different in a two-sample t test (P < 0.01). The horizontal line indicates a cutoff point of 2.1 where 11 of 13 AD subjects and 10 of 13 elderly subjects are correctly categorized, yielding a sensitivity of 85% and a specificity of 77%.
Similar articles
- Functional MRI assessment of task-induced deactivation of the default mode network in Alzheimer's disease and at-risk older individuals.
Pihlajamäki M, Sperling RA. Pihlajamäki M, et al. Behav Neurol. 2009;21(1):77-91. doi: 10.3233/BEN-2009-0231. Behav Neurol. 2009. PMID: 19847047 Free PMC article. - [Structural and functional neuronal connectivity in Alzheimer's disease: a combined DTI and fMRI study].
Soldner J, Meindl T, Koch W, Bokde AL, Reiser MF, Möller HJ, Bürger K, Hampel H, Teipel SJ. Soldner J, et al. Nervenarzt. 2012 Jul;83(7):878-87. doi: 10.1007/s00115-011-3326-3. Nervenarzt. 2012. PMID: 21713583 German. - Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease.
Zhou J, Greicius MD, Gennatas ED, Growdon ME, Jang JY, Rabinovici GD, Kramer JH, Weiner M, Miller BL, Seeley WW. Zhou J, et al. Brain. 2010 May;133(Pt 5):1352-67. doi: 10.1093/brain/awq075. Epub 2010 Apr 21. Brain. 2010. PMID: 20410145 Free PMC article. - Impact of Alzheimer's disease on the functional connectivity of spontaneous brain activity.
Sorg C, Riedl V, Perneczky R, Kurz A, Wohlschläger AM. Sorg C, et al. Curr Alzheimer Res. 2009 Dec;6(6):541-53. doi: 10.2174/156720509790147106. Curr Alzheimer Res. 2009. PMID: 19747154 Review. - Dementia and the default mode.
Beason-Held LL. Beason-Held LL. Curr Alzheimer Res. 2011 Jun;8(4):361-5. doi: 10.2174/156720511795745294. Curr Alzheimer Res. 2011. PMID: 21222595 Free PMC article. Review.
Cited by
- TMS as a Tool for Examining Cognitive Processing.
Nevler N, Ash EL. Nevler N, et al. Curr Neurol Neurosci Rep. 2015 Aug;15(8):52. doi: 10.1007/s11910-015-0575-8. Curr Neurol Neurosci Rep. 2015. PMID: 26092315 Review. - Brain network local interconnectivity loss in aging APOE-4 allele carriers.
Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, Bookheimer SY. Brown JA, et al. Proc Natl Acad Sci U S A. 2011 Dec 20;108(51):20760-5. doi: 10.1073/pnas.1109038108. Epub 2011 Nov 21. Proc Natl Acad Sci U S A. 2011. PMID: 22106308 Free PMC article. - Using regional homogeneity to reveal altered spontaneous activity in patients with mild cognitive impairment.
Wang Y, Zhao X, Xu S, Yu L, Wang L, Song M, Yang L, Wang X. Wang Y, et al. Biomed Res Int. 2015;2015:807093. doi: 10.1155/2015/807093. Epub 2015 Feb 9. Biomed Res Int. 2015. PMID: 25738156 Free PMC article. - Functional Connectivity in MRI Is Driven by Spontaneous BOLD Events.
Allan TW, Francis ST, Caballero-Gaudes C, Morris PG, Liddle EB, Liddle PF, Brookes MJ, Gowland PA. Allan TW, et al. PLoS One. 2015 Apr 29;10(4):e0124577. doi: 10.1371/journal.pone.0124577. eCollection 2015. PLoS One. 2015. PMID: 25922945 Free PMC article. - Structural architecture supports functional organization in the human aging brain at a regionwise and network level.
Zimmermann J, Ritter P, Shen K, Rothmeier S, Schirner M, McIntosh AR. Zimmermann J, et al. Hum Brain Mapp. 2016 Jul;37(7):2645-61. doi: 10.1002/hbm.23200. Epub 2016 Apr 4. Hum Brain Mapp. 2016. PMID: 27041212 Free PMC article.
References
- Minoshima, S., Giordani, B., Berent, S., Frey, K. A., Foster, N. L. & Kuhl, D. E. (1997) Ann. Neurol. 42, 85-94. - PubMed
- Johnson, K. A., Jones, K., Holman, B. L., Becker, J. A., Spiers, P. A., Satlin, A. & Albert, M. S. (1998) Neurology 50, 1563-1571. - PubMed
- Matsuda, H. (2001) Ann. Nucl. Med. 15, 85-92. - PubMed
- Reiman, E. M., Caselli, R. J., Yun, L. S., Chen, K., Bandy, D., Minoshima, S., Thibodeau, S. N. & Osborne, D. (1996) N. Engl. J. Med. 334, 752-758. - PubMed
Publication types
MeSH terms
Grants and funding
- T32 MH019938/MH/NIMH NIH HHS/United States
- MH01142/MH/NIMH NIH HHS/United States
- R01 HD031715/HD/NICHD NIH HHS/United States
- MH19938/MH/NIMH NIH HHS/United States
- K25 HD040761/HD/NICHD NIH HHS/United States
- HD31715/HD/NICHD NIH HHS/United States
- MH62430/MH/NIMH NIH HHS/United States
- HD40761/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases