A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier - PubMed (original) (raw)
Comparative Study
. 2004 May 5;23(9):1977-86.
doi: 10.1038/sj.emboj.7600205. Epub 2004 Apr 8.
Affiliations
- PMID: 15071506
- PMCID: PMC404325
- DOI: 10.1038/sj.emboj.7600205
Comparative Study
A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier
Masaaki Komatsu et al. EMBO J. 2004.
Abstract
Several studies have addressed the importance of various ubiquitin-like (UBL) post-translational modifiers. These UBLs are covalently linked to most, if not all, target protein(s) through an enzymatic cascade analogous to ubiquitylation, consisting of E1 (activating), E2 (conjugating), and E3 (ligating) enzymes. In this report, we describe the identification of a novel ubiquitin-fold modifier 1 (Ufm1) with a molecular mass of 9.1 kDa, displaying apparently similar tertiary structure, although lacking obvious sequence identity, to ubiquitin. Ufm1 is first cleaved at the C-terminus to expose its conserved Gly residue. This Gly residue is essential for its subsequent conjugating reactions. The C-terminally processed Ufm1 is activated by a novel E1-like enzyme, Uba5, by forming a high-energy thioester bond. Activated Ufm1 is then transferred to its cognate E2-like enzyme, Ufc1, in a similar thioester linkage. Ufm1 forms several complexes in HEK293 cells and mouse tissues, revealing that it conjugates to the target proteins. Ufm1, Uba5, and Ufc1 are all conserved in metazoa and plants but not in yeast, suggesting its potential roles in various multicellular organisms.
Figures
Figure 1
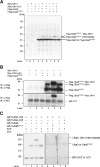
Uba5, a novel E1-like enzyme. (A) Schematic representation of Uba1 and Uba5 in H. sapiens. Uba1 is divided into several domains, including I, Ib, II, III, and IV boxes, which are conserved in other E1-like enzymes, and other regions without obvious similarity, described previously (Komatsu et al, 2001). Note that Uba5 is of a relatively small size and includes the box I and two other parts. The box I region of Uba1 (amino acids 459–611) has 48.4% similarity and 22.3% identity to amino acids 72–229 of Uba5, which includes the conserved ATP-binding motif (G_X_G_XX_G). The sequence of Uba5 is available from GenBanK™ under the accession number AK026904. hs, H. sapiens; ce, C. elegans; dm, D. melanogaster; at, A. thaliana. (B) Sequence alignment of hsUba5 and its homologs of other species (dm, NM_132494; ce, NM_058847; at, NM_100414). The amino-acid sequence of hsUba5 is compared by the ClustalW program. Asterisks, identical amino acids; single and double dots, weakly and strongly similar amino acids, respectively, determined by the criteria of ClustalW program. Open box indicates an ATP-binding motif. The putative active site Cys residue is boxed in black. The metal-binding motif is underlined. (C) Identification of the intermediate linked to Uba5 in HEK293 cells. Both Uba5 and Uba5C250S, in which the predicted active site Cys positioned at 250 was changed to Ser by site-directed mutagenesis, were tagged with Flag peptide at N-terminus, resulting in Flag-Uba5 and Flag-Uba5C250S, respectively. Each Flag-Uba5 and Flag-Uba5C250S was expressed in HEK293 cells. The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody.
Figure 2
Ufm1, a novel ubiquitin-fold molecule. (A) Sequence alignment of hsUfm1 and its homologs. The sequence of hsUfm1 is available from GenBanK™ under the accession number BC005193 (dm, a coding region of dmUfm1 was found from D. melanogaster genomic sequence; ce, NM_066304; at, NM_106420). The homology analysis was performed as described in Figure 1B. The C-terminal conserved Gly residue is boxed in black. (B) Sequence alignment of hsUbiquitin with hsUfm1. The homology analysis was performed as described in Figure 1B. The C-terminal conserved Gly residue is boxed in black. (C) Structural ribbon of hsUbiquitin and predicted structural ribbon of hsUfm1. α-Helices and β-strands are shown in green and yellow, respectively. The homology model of hsUfm1 was created from the C. elegans Ufm1 structure (Cort et al, 2002) by using MOE program (2003.02; Chemical Computing Group Inc., Montreal, Quebec, Canada). (D) Schematic representation of mammalian expression plasmids for Ufm1 and the derivative mutants. Flag epitope tags at the N-terminus, HA epitope tags at the C-terminus, and putative cleavage site Gly83 residue (vertical dotted lines) are indicated. To construct Ufm1G83A, a single point mutation was introduced into Ufm1, which led to an amino-acid substitution from Gly to Ala at position 83. To construct Ufm1ΔC2, the two C-terminal residues were deleted by PCR. Ufm1ΔC2G83A was also produced by site-directed mutagenesis of Ufm1ΔC2. The ΔC2 mutants were tagged with the Flag epitopes at N-terminus. (E) ProUfm1 processing. HEK293 cells were transfected with Flag-Ufm1-HA, Flag-Ufm1G83A-HA, Flag-Ufm1ΔC2, or Flag-Ufm1ΔC2G83A. The cell lysates were subjected to SDS–PAGE and analyzed by immunoblots with anti-Flag and anti-HA antibodies. ProUfm1 and mature Ufm1 are indicated on the left. The numbers at the top from I to IV are similar to those in (D).
Figure 3
Demonstration that Uba5 is an Ufm1-activating enzyme. (A) Immunoblotting analysis. Each Myc-tagged Ufm1 (Myc-Ufm1) and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Uba5 (lanes 5 and 6, respectively) or Flag-Uba5C250S (lanes 8 and 9, respectively). Each Flag-Uba5 and Flag-Uba5C250S was also expressed alone (lanes 4 and 7, respectively). The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bands corresponding to Flag-Uba5, Flag-Uba5C250S, and Flag-Uba5C250S intermediates are indicated on the right. (B) Immunoblotting analysis after immunoprecipitation. Each Myc-Ufm1 and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Uba5C250S (lanes 5 and 6, respectively). Flag-Uba5C250S was also expressed alone (lane 4). The cell lysates were immunoprecipitated with anti-Flag antibody. The resulting immunoprecipitates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag and anti-Myc antibodies. The bands corresponding to Flag-Uba5C250S, Flag-Uba5C250S–endogenous Ufm1, and Flag-Uba5C250S–Myc-Ufm1 intermediates are indicated. (C) In vitro activating assay of Ufm1 by Uba5. Purified recombinant GST-Ufm1ΔC2 (2 μg) (lanes 1–7) was incubated for 30 min at 25°C with some of the following: 2 μg of purified recombinant GST-Uba5 (lanes 2–5, 7, and 8), GST-Uba5C250A (lane 6), and 5 mM ATP (lanes 1 and 3–8). Lane 8 was conducted similar to lane 7, except that GST-Ufm1ΔC3 was used instead of GST-Ufm1ΔC2. Reactions were then incubated with SDS loading buffer lacking reducing agent (lanes 1–3 and 5–8) or containing 100 mM DTT (lane 4). The presence or absence of various components is indicated above the lanes. The bands corresponding to free GST-Uba5, GST-Uba5C250A, GST-Ufm1ΔC2 (mature Ufm1), GST-Ufm1ΔC3, and GST-Uba5–GST-Ufm1ΔC2 thioester product are indicated on the right.
Figure 4
Ufc1, a novel E2-like enzyme. (A) Sequence alignment of hsUfc1 and its homologs. The sequence of Ufc1 is available from GenBanK™ under the accession number BC005187 (dm, NM_137230; ce, NM_066654; at, BT001180). The homology analysis was performed as described in Figure 1B. The putative active site Cys residue is boxed in black. (B) Immunoblotting analysis. Each Myc-tagged Ufm1 (Myc-Ufm1) and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Ufc1 (lanes 5 and 6, respectively) or Flag-Ufc1C116S (lanes 8 and 9, respectively). Each Flag-Ufc1 and Flag-Ufc1C116S was also expressed alone (lanes 4 and 7, respectively). The cell lysates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bands corresponding to Flag-Ufc1, Flag-Ufc1C116S, and Flag-Ufc1C116S intermediates are indicated on the right. (C) Immunoblotting analysis after immunoprecipitation. Each Myc-Ufm1 and Myc-Ufm1ΔC3 was expressed alone (lanes 2 and 3, respectively), and coexpressed with Flag-Ufc1C116S (lanes 5 and 6, respectively). Flag-Ufc1C116S was also expressed alone (lane 4). The cell lysates were immunoprecipitated with anti-Flag antibody. The resulting immunoprecipitates were subjected to SDS–PAGE and analyzed by immunoblots with anti-Flag and anti-Myc antibodies. The bands corresponding to Flag-Ufc1C116S, Flag-Ufc1C116S–endogenous Ufm1, and Flag-Ufc1C116S–Myc-Ufm1 intermediates are indicated. (D) In vitro thioester bond formation assay of Ufm1 by Ufc1. Purified recombinant GST-Ufm1ΔC2 (2 μg) (lanes 1–8) was incubated for 30 min at 25°C with the following: purified recombinant GST-Uba5 (0.2 μg) (lanes 2–9), GST-Ufc1 (2 μg) (lanes 3–6, 8, and 9), GST-Ufc1C116S (2 μg) (lane 7), and 5 mM ATP (lanes 1, 2, and 4–9). Lane 9 was conducted similar to lane 8, except that GST-Ufm1ΔC3 was used instead of GST-Ufm1ΔC2. Reactions were then incubated with SDS loading buffer lacking reducing agent (lanes 1–4 and 6–9) or containing 100 mM DTT (lane 5). The presence or absence of various components is indicated above the lanes. The bands corresponding to free GST-Ufm1ΔC2 (mature Ufm1), GST-Ufm1ΔC3, GST-Uba5, GST-Ufc1, GST-Ufc1C116S, and GST-Ufc1–GST-Ufm1ΔC2 thioester product are indicated on the right.
Figure 5
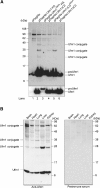
Formation of a covalent protein conjugate(s) with Ufm1 in HEK293 cells and mouse tissues. (A) Ufm1 conjugates in human HEK293 cells. HEK293 cells were transfected with FlagHis-Ufm1-HA, FlagHis-Ufm1G83A-HA, FlagHis-Ufm1ΔC2, FlagHis-Ufm1ΔC2G83A, or FlagHis-Ufm1ΔC3 expression plasmids. These cells were lysed under denaturing conditions, and the lysates were precipitated with Ni2+ beads. The precipitates were subjected to SDS–PAGE and analyzed by immunoblotting with anti-Flag antibody. The bottom panel shows the short exposure of the upper panel. The bands corresponding to mature Ufm1, proUfm, and Ufm1 conjugates are indicated on the right. (B) Ufm1 conjugates in various mouse tissues. Homogenates from mouse tissues as indicated were prepared and subjected to SDS–PAGE and analyzed by immunoblotting with anti-Ufm1 serum (left panel) or preimmune serum (right panel). The bands corresponding to Ufm1 and conjugates between Ufm1 and target proteins are indicated on the left.
Figure 6
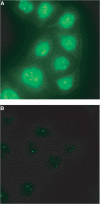
Intracellular distribution of Ufm1 in HeLa cells. (A) HeLa cells were seeded on coverslips 24 h before fixation for immunostaining. Ufm1 was detected with anti-Ufm1 serum and visualized with Alexa 488 nm anti-rabbit antibody. (B) Immunocytochemical analysis was conducted as for (A), except that preimmune serum was used. Cells were observed using a fluorescence microscope. Magnification, × 400.
Similar articles
- Structural and Functional Analysis of a Novel Interaction Motif within UFM1-activating Enzyme 5 (UBA5) Required for Binding to Ubiquitin-like Proteins and Ufmylation.
Habisov S, Huber J, Ichimura Y, Akutsu M, Rogova N, Loehr F, McEwan DG, Johansen T, Dikic I, Doetsch V, Komatsu M, Rogov VV, Kirkin V. Habisov S, et al. J Biol Chem. 2016 Apr 22;291(17):9025-41. doi: 10.1074/jbc.M116.715474. Epub 2016 Feb 29. J Biol Chem. 2016. PMID: 26929408 Free PMC article. - Mechanistic study of Uba5 enzyme and the Ufm1 conjugation pathway.
Gavin JM, Hoar K, Xu Q, Ma J, Lin Y, Chen J, Chen W, Bruzzese FJ, Harrison S, Mallender WD, Bump NJ, Sintchak MD, Bence NF, Li P, Dick LR, Gould AE, Chen JJ. Gavin JM, et al. J Biol Chem. 2014 Aug 15;289(33):22648-22658. doi: 10.1074/jbc.M114.573972. Epub 2014 Jun 25. J Biol Chem. 2014. PMID: 24966333 Free PMC article. - NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
Liu G, Forouhar F, Eletsky A, Atreya HS, Aramini JM, Xiao R, Huang YJ, Abashidze M, Seetharaman J, Liu J, Rost B, Acton T, Montelione GT, Hunt JF, Szyperski T. Liu G, et al. J Struct Funct Genomics. 2009 Apr;10(2):127-36. doi: 10.1007/s10969-008-9054-7. Epub 2008 Dec 20. J Struct Funct Genomics. 2009. PMID: 19101823 Free PMC article. - UFMylation: A Unique & Fashionable Modification for Life.
Wei Y, Xu X. Wei Y, et al. Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review. - Decrypting UFMylation: How Proteins Are Modified with UFM1.
Banerjee S, Kumar M, Wiener R. Banerjee S, et al. Biomolecules. 2020 Oct 14;10(10):1442. doi: 10.3390/biom10101442. Biomolecules. 2020. PMID: 33066455 Free PMC article. Review.
Cited by
- The Four Homeostasis Knights: In Balance upon Post-Translational Modifications.
Pieroni S, Castelli M, Piobbico D, Ferracchiato S, Scopetti D, Di-Iacovo N, Della-Fazia MA, Servillo G. Pieroni S, et al. Int J Mol Sci. 2022 Nov 21;23(22):14480. doi: 10.3390/ijms232214480. Int J Mol Sci. 2022. PMID: 36430960 Free PMC article. Review. - Ribosomal protein RPL26 is the principal target of UFMylation.
Walczak CP, Leto DE, Zhang L, Riepe C, Muller RY, DaRosa PA, Ingolia NT, Elias JE, Kopito RR. Walczak CP, et al. Proc Natl Acad Sci U S A. 2019 Jan 22;116(4):1299-1308. doi: 10.1073/pnas.1816202116. Epub 2019 Jan 9. Proc Natl Acad Sci U S A. 2019. PMID: 30626644 Free PMC article. - Bio-Guided Fractionation of Ethanol Extract of Leaves of Esenbeckia alata Kunt (Rutaceae) Led to the Isolation of Two Cytotoxic Quinoline Alkaloids: Evidence of Selectivity Against Leukemia Cells.
Álvarez-Caballero JM, Cuca-Suárez LE, Coy-Barrera E. Álvarez-Caballero JM, et al. Biomolecules. 2019 Oct 8;9(10):585. doi: 10.3390/biom9100585. Biomolecules. 2019. PMID: 31597257 Free PMC article. - Modification of ERα by UFM1 Increases Its Stability and Transactivity for Breast Cancer Development.
Yoo HM, Park JH, Kim JY, Chung CH. Yoo HM, et al. Mol Cells. 2022 Jun 30;45(6):425-434. doi: 10.14348/molcells.2022.0029. Mol Cells. 2022. PMID: 35680375 Free PMC article. - DDRGK1 is required for the proper development and maintenance of the growth plate cartilage.
Weisz-Hubshman M, Egunsula AT, Dawson B, Castellon A, Jiang MM, Chen-Evenson Y, Zhiyin Y, Lee B, Bae Y. Weisz-Hubshman M, et al. Hum Mol Genet. 2022 Aug 23;31(16):2820-2830. doi: 10.1093/hmg/ddac078. Hum Mol Genet. 2022. PMID: 35377455 Free PMC article.
References
- Cort JR, Chiang Y, Zheng D, Montelione GT, Kennedy MA (2002) NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold. Proteins 48: 733–736 - PubMed
- Furukawa K, Mizushima N, Noda T, Ohsumi Y (2000) A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem 275: 7462–7465 - PubMed
- Glickman MH, Ciechanover A (2002) The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases