Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome - PubMed (original) (raw)
Case Reports
Novel PMS2 pseudogenes can conceal recessive mutations causing a distinctive childhood cancer syndrome
Michel De Vos et al. Am J Hum Genet. 2004 May.
Abstract
We investigated a family with an autosomal recessive syndrome of cafe-au-lait patches and childhood malignancy, notably supratentorial primitive neuroectodermal tumor. There was no cancer predisposition in heterozygotes; nor was there bowel cancer in any individual. However, autozygosity mapping indicated linkage to a region of 7p22 surrounding the PMS2 mismatch-repair gene. Sequencing of genomic PCR products initially failed to identify a PMS2 mutation. Genome searches then revealed a previously unrecognized PMS2 pseudogene, corresponding to exons 9-15, within a 100-kb inverted duplication situated 600 kb centromeric from PMS2 itself. This information allowed a redesigned sequence analysis, identifying a homozygous mutation (R802X) in PMS2 exon 14. Furthermore, in the family with Turcot syndrome, in which the first inherited PMS2 mutation (R134X) was described, a further truncating mutation was identified on the other allele, in exon 13. Further whole-genome analysis shows that the complexity of PMS2 pseudogenes is greater than appreciated and may have hindered previous mutation studies. Several previously reported PMS2 polymorphisms are, in fact, pseudogene sequence variants. Although PMS2 mutations may be rare in colorectal cancer, they appear, for the most part, to behave as recessive traits. For technical reasons, their involvement in childhood cancer, particularly in primitive neuroectodermal tumor, may have been underestimated.
Figures
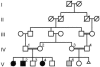
Figure 1
Pedigree diagram. Linkage analysis was performed by use of DNA from the eight numbered individuals (IV-1–IV-4, V-2–V-4, V-6). Gray shading of the square representing patient V-6 indicates that he has CALS but no tumor.
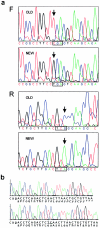
Figure 2
a, Mutation analysis of PMS2 exon 14 (ABI377). Forward (F) and reverse (R) strand sequences are shown of uncloned PCR products generated by use of the published primer pair (upper panels) or the redesigned _PMS2_-specific primers (lower panels). The mutated codon is boxed, and the arrows indicate the mutated nucleotide. b, Exon 13 mutation in the patient with Turcot syndrome (Hamilton et al. 1995) (MegaBace). PCR products were cloned to show both deleted (top) and normal alleles. The lowercase nucleotides are the end of intron 12. The deleted dinucleotide (within a 2-nt repeat) is underlined, and the resulting predicted novel C-terminus is italicized.
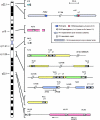
Figure 3
Distribution of PMS2 pseudogenes and related sequences on chromosome 7. Symbols are as defined in the key. The members of the pseudogene-associated repeat family (PPJ repeats) can be classified by sequence similarity into two main subgroups (indicated by white and gray block arrows) and two outlying members (turquoise). The type 1 members (gray) appear to be the younger of the two main PPJ subgroups, having (with the exception of the ψ_13_-associated PPJ element) shorter distance branches on the phylogeny tree. Larger blocks of different colors indicate different higher-order repeat elements. Those shown in yellow and green correspond, respectively, to the type A and type C repeat elements identified elsewhere in studies of the Williams syndrome deletion region (Valero et al. 2000). (Other large repeat blocks in this region that are not associated with PPJ elements are not shown.) Numbers indicate the approximate positions (in Mb) of each PPJ element on the draft genome sequence (build 33).
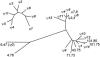
Figure 4
Phylogenetic tree illustrating the relationships between the different members of the family with the 23-kb PPJ element. The tree was generated by using the DNADIST algorithm of the PHYLIP package to analyze a ClustalW-generated alignment of all 20 copies of a ∼1.5-kb segment of the PPJ element. (Details are available on request.) Branch lengths, therefore, give an indication of degree of divergence. The pseudogene-associated PPJ elements are referred to by the corresponding identity (e.g., ψ_1_). Those not associated with a pseudogene are given a number representing their sequence coordinate on chromosome 7 (National Center for Biotechnology Information [NCBI] release 33). The inset shows a tree generated from a subset of these PPJ elements by use of the same algorithm, to show more clearly the interrelationships of the closely similar ψ_2_–ψ_8_ PPJ elements. The structure of this tree implies that the local duplications leading to these PPJ clusters (and, hence, associated pseudogene clusters) occurred after the large-scale (∼100-kb) duplications around the Williams syndrome deletion region.
Similar articles
- Polymorphisms in a pseudogene highly homologous to PMS2.
Chadwick RB, Meek JE, Prior TW, Peltomaki P, de La Chapelle A. Chadwick RB, et al. Hum Mutat. 2000 Dec;16(6):530. doi: 10.1002/1098-1004(200012)16:6<530::AID-HUMU15>3.0.CO;2-6. Hum Mutat. 2000. PMID: 11102987 - PMS2 mutations in childhood cancer.
De Vos M, Hayward BE, Charlton R, Taylor GR, Glaser AW, Picton S, Cole TR, Maher ER, McKeown CM, Mann JR, Yates JR, Baralle D, Rankin J, Bonthron DT, Sheridan E. De Vos M, et al. J Natl Cancer Inst. 2006 Mar 1;98(5):358-61. doi: 10.1093/jnci/djj073. J Natl Cancer Inst. 2006. PMID: 16507833 - Extensive gene conversion at the PMS2 DNA mismatch repair locus.
Hayward BE, De Vos M, Valleley EM, Charlton RS, Taylor GR, Sheridan E, Bonthron DT. Hayward BE, et al. Hum Mutat. 2007 May;28(5):424-30. doi: 10.1002/humu.20457. Hum Mutat. 2007. PMID: 17253626 - Phenotype associated with recessively inherited mutations in DNA mismatch repair (MMR) genes.
de Vos M, Hayward B, Bonthron DT, Sheridan E. de Vos M, et al. Biochem Soc Trans. 2005 Aug;33(Pt 4):718-20. doi: 10.1042/BST0330718. Biochem Soc Trans. 2005. PMID: 16042583 Review. - Paediatric intestinal cancer and polyposis due to bi-allelic PMS2 mutations: case series, review and follow-up guidelines.
Herkert JC, Niessen RC, Olderode-Berends MJ, Veenstra-Knol HE, Vos YJ, van der Klift HM, Scheenstra R, Tops CM, Karrenbeld A, Peters FT, Hofstra RM, Kleibeuker JH, Sijmons RH. Herkert JC, et al. Eur J Cancer. 2011 May;47(7):965-82. doi: 10.1016/j.ejca.2011.01.013. Epub 2011 Mar 4. Eur J Cancer. 2011. PMID: 21376568 Review.
Cited by
- PMS2 monoallelic mutation carriers: the known unknown.
Goodenberger ML, Thomas BC, Riegert-Johnson D, Boland CR, Plon SE, Clendenning M, Win AK, Senter L, Lipkin SM, Stadler ZK, Macrae FA, Lynch HT, Weitzel JN, de la Chapelle A, Syngal S, Lynch P, Parry S, Jenkins MA, Gallinger S, Holter S, Aronson M, Newcomb PA, Burnett T, Le Marchand L, Pichurin P, Hampel H, Terdiman JP, Lu KH, Thibodeau S, Lindor NM. Goodenberger ML, et al. Genet Med. 2016 Jan;18(1):13-9. doi: 10.1038/gim.2015.27. Epub 2015 Apr 9. Genet Med. 2016. PMID: 25856668 Free PMC article. Review. - Multifocal anaplastic astrocytoma in a patient with hereditary colorectal cancer, transcobalamin II deficiency, agenesis of the corpus callosum, mental retardation, and inherited PMS2 mutation.
Gururangan S, Frankel W, Broaddus R, Clendenning M, Senter L, McDonald M, Eastwood J, Reardon D, Vredenburgh J, Quinn J, Friedman HS. Gururangan S, et al. Neuro Oncol. 2008 Feb;10(1):93-7. doi: 10.1215/15228517-2007-037. Epub 2007 Nov 9. Neuro Oncol. 2008. PMID: 17993636 Free PMC article. - PMS2 mutations in childhood cancer.
Bonthron DT, Hayward BE, De Vos M, Sheridan E. Bonthron DT, et al. Gut. 2005 Dec;54(12):1821. doi: 10.1136/gut.2005.078816. Gut. 2005. PMID: 16284300 Free PMC article. No abstract available. - Human PMS2 gene family: origin, molecular evolution, and biological implications.
Shpakovskii DG, Shematorova EK, Shpakovskii GV. Shpakovskii DG, et al. Dokl Biochem Biophys. 2006 May-Jun;408:175-9. doi: 10.1134/s1607672906030185. Dokl Biochem Biophys. 2006. PMID: 16913423 No abstract available. - A sensitive and scalable microsatellite instability assay to diagnose constitutional mismatch repair deficiency by sequencing of peripheral blood leukocytes.
Gallon R, Mühlegger B, Wenzel SS, Sheth H, Hayes C, Aretz S, Dahan K, Foulkes W, Kratz CP, Ripperger T, Azizi AA, Baris Feldman H, Chong AL, Demirsoy U, Florkin B, Imschweiler T, Januszkiewicz-Lewandowska D, Lobitz S, Nathrath M, Pander HJ, Perez-Alonso V, Perne C, Ragab I, Rosenbaum T, Rueda D, Seidel MG, Suerink M, Taeubner J, Zimmermann SY, Zschocke J, Borthwick GM, Burn J, Jackson MS, Santibanez-Koref M, Wimmer K. Gallon R, et al. Hum Mutat. 2019 May;40(5):649-655. doi: 10.1002/humu.23721. Epub 2019 Mar 6. Hum Mutat. 2019. PMID: 30740824 Free PMC article.
References
Electronic-Database Information
- MegaBLAST, http://www.ncbi.nlm.nih.gov/genome/seq
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for retinoblastoma, Li-Fraumeni syndrome, SMARCB1, NF1, and Turcot syndrome)
- RepeatMasker, http://woody.embl-heidelberg.de/repeatmask
References
- Baptiste M, Nasca P, Metzger B, Field N, MacCubbin P, Greenwald P, Armbrustmacher V, Waldman J, Carlton K (1989) Neurofibromatosis and other disorders among children with CNS tumors and their families. Neurology 39:487–492 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases