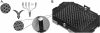Decoding randomly ordered DNA arrays - PubMed (original) (raw)
doi: 10.1101/gr.2255804. Epub 2004 Apr 12.
Semyon Kruglyak, Michael S Graige, Francisco Garcia, Bahram G Kermani, Chanfeng Zhao, Diping Che, Todd Dickinson, Eliza Wickham, Jim Bierle, Dennis Doucet, Monika Milewski, Robert Yang, Chris Siegmund, Juergen Haas, Lixin Zhou, Arnold Oliphant, Jian-Bing Fan, Steven Barnard, Mark S Chee
Affiliations
- PMID: 15078854
- PMCID: PMC479114
- DOI: 10.1101/gr.2255804
Decoding randomly ordered DNA arrays
Kevin L Gunderson et al. Genome Res. 2004 May.
Abstract
We have developed a simple and efficient algorithm to identify each member of a large collection of DNA-linked objects through the use of hybridization, and have applied it to the manufacture of randomly assembled arrays of beads in wells. Once the algorithm has been used to determine the identity of each bead, the microarray can be used in a wide variety of applications, including single nucleotide polymorphism genotyping and gene expression profiling. The algorithm requires only a few labels and several sequential hybridizations to identify thousands of different DNA sequences with great accuracy. We have decoded tens of thousands of arrays, each with 1520 sequences represented at approximately 30-fold redundancy by up to approximately 50,000 beads, with a median error rate of <1 x 10(-4) per bead. The approach makes use of error checking codes and provides, for the first time, a direct functional quality control of every element of each array that is manufactured. The algorithm can be applied to any spatially fixed collection of objects or molecules that are associated with specific DNA sequences.
Figures
Figure 1
Assembly of a random array. (A) Creation of a bead pool and assembly into ∼3-μm-diameter wells etched in optical fiber bundles. Once a bead pool is made, it is relatively straightforward to assemble and decode large numbers of arrays. Each array contains ∼50,000 beads distributed among 1520 bead types, so that each bead type is represented at ∼30-fold redundancy. Scanning electron micrographs are shown of an unassembled and an assembled array containing one bead per well. (B) Because individual arrays are only ∼1.4 mm in diameter, they can easily be arranged into a 96-array matrix, designed for parallel analysis of samples in standard microtiter plates.
Figure 2
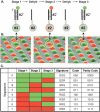
Decoding process. (A) The sequential hybridization process is illustrated for a single bead, of bead type 2. In stage 1, a complementary decoder hybridizes to the oligonucleotide probe that is attached to the bead (for details of the procedure, see Methods). The decoder is labeled with a fluorophore (green in stage 1, red in stage 2, and green in stage 3). The fluorescent signal is read by imaging the entire array. The array is then dehybridized, and the process is repeated for two more stages. (B) A scanning electron micrograph of an array of beads, artificially colored to represent three sequential hybridization stages. The images, taken collectively, reveal a combinatorial code for each bead. Note that the bead circled in yellow has the color signature GRG or code 010. (C) Colors, or states, are assigned to individual decoder sequences in each stage to produce a unique combination across stages. This signature, or code, identifies each bead type. As indicated in the parity code column, an extra decoding stage (data not shown) can be performed to provide an error checking parity bit. After three stages of decoding, all the beads are uniquely identified by their color.
Figure 3
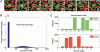
(A) Decoding images from eight sequential stages (numbered). Each image is a false-color composite of the FAM and Cy3 grayscale images from each stage. A small region (<0.2%) of a single array is shown. The circled bead is one of up to ∼4.8 million in a single 96-array matrix. It has the code 11012202. A total of 1728 images, each ∼5 Mb in size and containing ∼1.7 million pixels, are collected in order to decode an array matrix. (B) Histogram of bead intensities for a single stage. The low intensity peak includes beads in the OFF state and empty wells. The higher intensity peak includes beads in the ON state. (C) The eight-stage intensity profile of a typical individual bead (code = 02212110) in the FAM and Cy3 channels (∼100 counts of intensity are from camera DC offset).
Figure 4
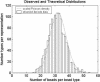
Bead representation histogram from a representative decoded array overlaid with a scaled Poisson density function. The loading of each array is a sampling of beads from a near infinite bead pool. Almost N ∼ 50,000 beads are sampled, and the probability, p, that a sampled bead belongs to a particular type is approximately one divided by the number of bead types. Because N is large, p is small, and the initial bead pool is near infinite, the number of beads from each bead type is well modeled by a Poisson distribution with mean Np. With 1520 bead types, we average >30 replicate beads per bead type, and the probability that any bead type has fewer than five replicate beads is extremely low (∼5.5 × 10-6).
Similar articles
- The distribution and deposition algorithm for multiple oligo nucleotide arrays.
Ning K, Leong HW. Ning K, et al. Genome Inform. 2006;17(2):89-99. Genome Inform. 2006. PMID: 17503382 - A multi-array multi-SNP genotyping algorithm for Affymetrix SNP microarrays.
Xiao Y, Segal MR, Yang YH, Yeh RF. Xiao Y, et al. Bioinformatics. 2007 Jun 15;23(12):1459-67. doi: 10.1093/bioinformatics/btm131. Epub 2007 Apr 25. Bioinformatics. 2007. PMID: 17459966 - Handling long targets and errors in sequencing by hybridization.
Halperin E, Halperin S, Hartman T, Shamir R. Halperin E, et al. J Comput Biol. 2003;10(3-4):483-97. doi: 10.1089/10665270360688138. J Comput Biol. 2003. PMID: 12935339 - High-throughput SNP genotyping on universal bead arrays.
Shen R, Fan JB, Campbell D, Chang W, Chen J, Doucet D, Yeakley J, Bibikova M, Wickham Garcia E, McBride C, Steemers F, Garcia F, Kermani BG, Gunderson K, Oliphant A. Shen R, et al. Mutat Res. 2005 Jun 3;573(1-2):70-82. doi: 10.1016/j.mrfmmm.2004.07.022. Mutat Res. 2005. PMID: 15829238 Review. - Fabrication of DNA microarray.
Dufva M. Dufva M. Methods Mol Biol. 2009;529:63-79. doi: 10.1007/978-1-59745-538-1_5. Methods Mol Biol. 2009. PMID: 19381969 Review.
Cited by
- Encoding microcarriers for biomedicine.
Wei X, Shang Y, Zhu Y, Gu Z, Zhang D. Wei X, et al. Smart Med. 2023 Feb 14;2(1):e20220009. doi: 10.1002/SMMD.20220009. eCollection 2023 Feb. Smart Med. 2023. PMID: 39188559 Free PMC article. Review. - Screening of Methyl-β-cyclodextrins as an Antifading Agent for Cyanine Dye-Labeled Streptavidin to Improve the Performance of Genotyping Chips.
Ma Y, Fan Y, Xu X, Li H, Liu R, Liu C. Ma Y, et al. ACS Omega. 2024 Jun 27;9(27):29491-29498. doi: 10.1021/acsomega.4c02099. eCollection 2024 Jul 9. ACS Omega. 2024. PMID: 39005797 Free PMC article. - High-definition spatial transcriptomics for in situ tissue profiling.
Vickovic S, Eraslan G, Salmén F, Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R, Bergenstråhle L, Navarro JF, Gould J, Griffin GK, Borg Å, Ronaghi M, Frisén J, Lundeberg J, Regev A, Ståhl PL. Vickovic S, et al. Nat Methods. 2019 Oct;16(10):987-990. doi: 10.1038/s41592-019-0548-y. Epub 2019 Sep 9. Nat Methods. 2019. PMID: 31501547 Free PMC article. - A trap-and-release integrated microfluidic system for dynamic microarray applications.
Tan WH, Takeuchi S. Tan WH, et al. Proc Natl Acad Sci U S A. 2007 Jan 23;104(4):1146-51. doi: 10.1073/pnas.0606625104. Epub 2007 Jan 16. Proc Natl Acad Sci U S A. 2007. PMID: 17227861 Free PMC article. - Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene.
Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Alarcón M, et al. Am J Hum Genet. 2008 Jan;82(1):150-9. doi: 10.1016/j.ajhg.2007.09.005. Am J Hum Genet. 2008. PMID: 18179893 Free PMC article.
References
- Alwine, J.C., Kemp, D.J., Parker, B.A., Reiser, J., Renart, J., Stark, G.R., and Wahl, G.M. 1979. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 68: 220-242. - PubMed
- Barker, D.L., Therault, G., Che, D., Dickinson, T., Shen, R., and Kain, R. 2003. Self-assembled random arrays: High-performance imaging and genomics applications on a high-density microarray platform. Proc. SPIE 4966: 1-11.
- Battaglia, C., Salani, G., Consolandi, C., Bernardi, L.R., and De Bellis, G. 2000. Analysis of DNA microarrays by non-destructive fluorescent staining using SYBR green II. Biotechniques 29: 78-81. - PubMed
- Braeckmans, K., De Smedt, S.C., Leblans, M., Pauwels, R., and Demeester, J. 2002. Encoding microcarriers: Present and future technologies. Nat. Rev. Drug Discov. 1: 447-456. - PubMed
- Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D.H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. 2000. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630-634. - PubMed
WEB SITE REFERENCES
- www.hapmap.org; International HapMap Project.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources