A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast - PubMed (original) (raw)
A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast
Shakil Ahmed et al. Mol Cell Biol. 2004 May.
Abstract
The conserved protein kinase Chk1 mediates cell cycle progression and consequently the ability of cells to survive when exposed to DNA damaging agents. Cells deficient in Chk1 are hypersensitive to such agents and enter mitosis in the presence of damaged DNA, whereas checkpoint-proficient cells delay mitotic entry to permit time for DNA repair. In a search for proteins that can improve the survival of Chk1-deficient cells exposed to DNA damage, we identified fission yeast Msc1, which is homologous to a mammalian protein that binds to the tumor suppressor Rb (RBP2). Msc1 and RBP2 each possess three PHD fingers, domains commonly found in proteins that influence the structure of chromatin. Msc1 is chromatin associated and coprecipitates a histone deacetylase activity, a property that requires the PHD fingers. Cells lacking Msc1 have a dramatically altered histone acetylation pattern, exhibit a 20-fold increase in global acetylation of histone H3 tails, and are readily killed by trichostatin A, an inhibitor of histone deacetylases. We postulate that Msc1 plays an important role in regulating chromatin structure and that this function modulates the cellular response to DNA damage.
Figures
FIG. 1.
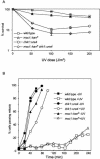
Isolation of Msc1 as a multicopy suppressor of loss of checkpoint function. (A) Screening used to isolate the multicopy suppressors of loss of checkpoint function (see text for details). (B) chk1::ura cdc17-K42 strains transformed with either an empty vector or a plasmid with a genomic copy of msc1 (p_msc1_) were grown to mid-log phase in liquid culture. Tenfold serial dilutions were made, and aliquots were spotted on plates. Plates were incubated at 25 or 32°C for 3 days. (C) Analysis of Chk1 phosphorylation in strains with defective DNA ligase activity of cdc17-K42 at 32°C. A cdc17-K42 strain with an integrated HA-tagged chk1 allele was transformed with either empty vector (lanes 1 and 2) or with pMsc1 (lanes 3 and 4). Strains were grown at 25°C to mid-log phase and then shifted to 32°C for 6 h. Protein was extracted by glass bead lysis, separated on an sodium dodecyl sulfate-polyacrylamide gel, transferred to nitrocellulose membrane, and blotted with antibody to the HA tag to detect the unphosphorylated and phosphorylated forms of Chk1. (D) A chk1::ura4 deletion strain was transformed with empty vector or plasmids containing genomic copies of msc1 or chk1. Strains were grown in liquid culture to mid-log phase, and 1,000 cells were plated and exposed to the indicated doses of UV light. All plates were incubated at 30°C for 3 days. The percentages of surviving colonies relative to those seen with unirradiated control plates were determined. Values shown are the averages of three independent experiments.
FIG. 2.
Loss of msc1 function compounds the sensitivity to UV light of a _chk1_-deficient strain. (A) The indicated strains were grown in rich medium to mid-log phase, and 1,000 cells per plate were exposed (or not exposed) to the indicated doses of UV light. Survival after 3 days on plates was determined as described in the legend to Fig. 1D. (B) The UV sensitivity of a strain lacking msc1 is not due to a compromised checkpoint. The indicated strains (each having a cdc25-22 mutant allele) were synchronized in G2 by incubation at 36.5°C, exposed to UV light, and released to permissive temperature to monitor passage through mitosis as described in Materials and Methods.
FIG. 3.
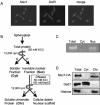
Domain architecture of Msc1. The domain architecture of Msc1 has the same motif arrangement as that of RBP2 (GenBank accession number NP_005047) and PLU-1 (GenBank accession number CAB63108). Msc1 was investigated using the GenBank database and the CDART available at the National Center for Biotechnology Information (
http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd = rps
) a.a., amino acids.
FIG. 4.
Msc1 is a chromatin-associated protein. (A) Nuclear localization of Msc1. Cells with an integrated allele of HA-tagged msc1 were grown to mid-log phase and fixed with glutaraldehyde, and immunofluorescence assays were performed using anti-HA antibody. (B) Schematic representation of chromatin fractionation assay. (C) DNA was isolated (as described in Materials and Methods) from the indicated fractions, run on an agarose gel, and stained with ethidium bromide. (D) Protein samples from the indicated fractions were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose. Western blot analysis was performed using anti-HA antibody to detect HA-tagged Msc1 (Msc1-HA), anti-Ded1 antibody (Ded1), or antibody to histone H4.
FIG. 5.
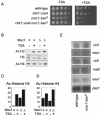
Deletion of msc1 affects cellular sensitivity to TSA and alters the state of histone acetylation in vivo. (A) An msc1::kanR deletion strain is TSA sensitive. Tenfold serial dilutions of the indicated strains were spotted on rich medium (−TSA) or rich medium containing 25 μg of TSA/ml (+TSA) and incubated at 30°C for 3 days. (B) Deletion of Msc1 results in hyperacetylation of histone H3. Histones were isolated from wild-type cells (Msc1 +) or an msc1::kanR deletion strain (Msc1 Δ). Histones (25 μg) were loaded on a 15% polyacrylamide gel and transferred to nylon membrane. Acetylated (Ac) histones were detected using antibody that recognizes histone H3 diacetylated on lysines 9 and 14 (upper panel), total histone H3 (middle panel), or histone H4 tetra-acetylated on lysines 5, 8, 12, and 16 (lower panel). (C and D) The data presented in panel B was quantitated using ImageQuant software normalized with histone H3 values and plotted to convey the relative amounts of acetylation in the different strains. (E) Northern blot analysis of genes affecting histone acetylation. RNA was isolated from wild-type and msc1::kanR cells and probed for the expression level of the indicated genes. The clr3, hda1, and clr6 genes encode HDAC. The mst2 and esa1 genes encode putative HAT, as suggested by sequence similarity to genes in other organisms. The ded1 gene, encoding a DEAD-box helicase involved in translation initiation, was used as a loading control.
FIG. 6.
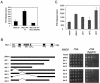
Msc1 associates with a HDAC. Msc1 coprecipitates a HDAC activity. (A) A strain having HA-tagged Msc1 was grown to mid-log phase, and Msc1 was immunoprecipitated (IP) as described in Materials and Methods. Mock or Msc1 immunoprecipitated samples were incubated with labeled histone H4 peptide for 24 h at room temperature. One set of Msc1 immunoprecipitates was incubated in the presence of sodium butyrate (Sod. Butyrate). Released 3H was counted using a scintillation counter. The values shown represent the averages of three assays, and the error bars represent the standard deviations of the data. (B) An msc1::kanr deletion strain was transformed with the indicated plasmids. Transformants were grown to mid-log phase, and 10-fold serial dilutions of the indicated strains were spotted on EMM-leu medium in the absence (−TSA) or presence (+TSA) of 5 μg of TSA/ml. Plates were incubated at 30°C for 4 days. (C) Strains harboring plasmids expressing deletion constructs of HA-tagged Msc1 were grown to mid-log phase, and Msc1 was immunoprecipitated and assayed for HDAC activity as described for panel A.
Similar articles
- Kinetochore Components Required for Centromeric Chromatin Assembly Are Impacted by Msc1 in Schizosaccharomyces pombe.
Gao C, Langbein L, Kamal F, George AA, Walworth NC. Gao C, et al. Genetics. 2017 Oct;207(2):559-569. doi: 10.1534/genetics.117.300183. Epub 2017 Aug 21. Genetics. 2017. PMID: 28827290 Free PMC article. - Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe.
Ahmed S, Dul B, Qiu X, Walworth NC. Ahmed S, et al. Genetics. 2007 Nov;177(3):1487-97. doi: 10.1534/genetics.107.078691. Epub 2007 Oct 18. Genetics. 2007. PMID: 17947424 Free PMC article. - The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity.
Dul BE, Walworth NC. Dul BE, et al. J Biol Chem. 2007 Jun 22;282(25):18397-18406. doi: 10.1074/jbc.M700729200. Epub 2007 Apr 24. J Biol Chem. 2007. PMID: 17456468 - Histone acetylation and the cell-cycle in cancer.
Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Wang C, et al. Front Biosci. 2001 Apr 1;6:D610-29. doi: 10.2741/1wang1. Front Biosci. 2001. PMID: 11282573 Review. - Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function.
Yoshida M, Horinouchi S, Beppu T. Yoshida M, et al. Bioessays. 1995 May;17(5):423-30. doi: 10.1002/bies.950170510. Bioessays. 1995. PMID: 7786288 Review.
Cited by
- Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease.
Cloos PA, Christensen J, Agger K, Helin K. Cloos PA, et al. Genes Dev. 2008 May 1;22(9):1115-40. doi: 10.1101/gad.1652908. Genes Dev. 2008. PMID: 18451103 Free PMC article. Review. - Polycomb-dependent nucleolus localization of Jumonji/Jarid2 during Drosophila spermatogenesis.
Goto M, Toda N, Shimaji K, Suong DN, Vo N, Kimura H, Yoshida H, Inoue YH, Yamaguchi M. Goto M, et al. Spermatogenesis. 2016 Sep 3;6(3):e1232023. doi: 10.1080/21565562.2016.1232023. eCollection 2016. Spermatogenesis. 2016. PMID: 28144496 Free PMC article. - Kinetochore Components Required for Centromeric Chromatin Assembly Are Impacted by Msc1 in Schizosaccharomyces pombe.
Gao C, Langbein L, Kamal F, George AA, Walworth NC. Gao C, et al. Genetics. 2017 Oct;207(2):559-569. doi: 10.1534/genetics.117.300183. Epub 2017 Aug 21. Genetics. 2017. PMID: 28827290 Free PMC article. - Msc1 links dynamic Swi6/HP1 binding to cell fate determination.
Lawrence RJ, Volpe TA. Lawrence RJ, et al. Proc Natl Acad Sci U S A. 2009 Jan 27;106(4):1163-8. doi: 10.1073/pnas.0811161106. Epub 2009 Jan 21. Proc Natl Acad Sci U S A. 2009. PMID: 19164572 Free PMC article. - Identification of histone demethylases in Saccharomyces cerevisiae.
Tu S, Bulloch EM, Yang L, Ren C, Huang WC, Hsu PH, Chen CH, Liao CL, Yu HM, Lo WS, Freitas MA, Tsai MD. Tu S, et al. J Biol Chem. 2007 May 11;282(19):14262-71. doi: 10.1074/jbc.M609900200. Epub 2007 Mar 16. J Biol Chem. 2007. PMID: 17369256 Free PMC article.
References
- Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. - PubMed
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. - PubMed
- Balciunas, D., and H. Ronne. 2000. Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25:274-276. - PubMed
- Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous