Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene - PubMed (original) (raw)
Identification of nucleophosmin as an NF-kappaB co-activator for the induction of the human SOD2 gene
Sanjit K Dhar et al. J Biol Chem. 2004.
Abstract
Manganese superoxide dismutase (MnSOD) is an antioxidant enzyme essential for the survival of life. We have reported that NF-kappaB is essential but not sufficient for the synergistic induction of MnSOD by phorbol 12-myristate 13-acetate and cytokines. To further identify transcription factors and co-activators that participate in the induction of MnSOD, we used NF-kappaB affinity chromatography to isolate potential NF-kappaB interacting proteins. Proteins eluted from the NF-kappaB affinity column were subjected to proteomic analysis and verified by Western analysis. Nucleophosmin (NPM), a nucleolar phosphoprotein, is the most abundant single protein identified. Co-immunoprecipitation studies suggest a physical interaction between NPM and NF-kappaB proteins. To verify the role of NPM on MnSOD gene transcription, cells were transfected with constructs expressing NPM in sense or antisense orientation as well as interference RNA. The results indicate that an increase NPM expression leads to increased MnSOD gene transcription in a dose-dependent manner. Consistent with this, expression of small interfering RNA for NPM leads to inhibition of MnSOD gene transcription but does not have any effect on the expression of interleukin-8, suggesting that the effect of NPM is selective. These results identify NPM as a partner of the NF-kappaB transcription complex in the induction of MnSOD by phorbol 12-myristate 13-acetate and cytokines.
Figures
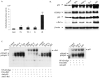
Fig. 1. Induction of MnSOD gene transcription after treatment of PMA and cytokines
A, subconfluent HepG2 cells were transiently transfected with MnSOD promoter- and enhancer-driven pGL3 reporter vectors along with Renilla (pRL-TK) luciferase vector as an internal control. Eight hours after transfections, cells were washed with 1x PBS and incubated at 37 °C for 24 h. At 12 h after treatment of PMA and cytokines, cell lysates were collected. Reporter activity was determined as a measure of MnSOD gene transcription by relative chemiluminescent light units. B, nuclear extracts were collected from HepG2 cells and the levels of p50, p65, CEBP/β, and c-Rel in HepG2 cells after treatment of PMA or cytokines or in combination for 4 h was determined by Western blotting analysis. Equal amounts of nuclear extracts (50 _μ_g) were subjected to 10% SDS-polyacrylamide gel electrophoresis, and Western analysis was performed using antibodies specific to p50, p65, CEBP/β, and c-Rel. The same membrane was re-probed with a _β_-actin antibody used as the loading control. C, NF-_κ_B DNA binding reactions were carried out as described under “Experimental Procedures.” For supershift experiments nuclear extracts were preincubated with 1 _μ_g of an antibody specific to p50, p65, and c-Rel prior to the binding reaction. DNA binding complexes and supershift complexes are indicated by the arrows. Each data point represents the mean of three independent experiments mean ± S.D. Significant difference from control: *, p < 0.05, and **, p < 0.01.
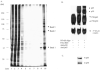
Fig. 2. Affinity purified DNA-binding proteins
A, purification of NF-_κ_B DNA-binding proteins by NF-_κ_B oligonucleotide DNA affinity chromatography. Various fractions obtained from the NF-_κ_B affinity column were subjected to 10% SDS-polyacrylamide gel electrophoresis and thereafter visualized by silver staining. Lane 1 represents the crude nuclear extract; lanes 2 and 3 represent the first flow-through and first wash flow-through with 800 mM KCl containing binding buffer, respectively. Lanes 4–8 represent the wash flow-through with 1x binding buffer. Lane 9 is 150 mM KCl in 1x binding buffer wash flow-through, and lane 10 is 450 mM KCl in 1x binding buffer elute. B, EMSA of affinity purified NF-_κ_B DNA-binding protein complexes. For the supershift assay, anti-p50 or anti-p65 antibodies of 1 _μ_g were preincubated with the EMSA reaction mixture, and the reaction was performed as described under “Experimental Procedures.” DNA binding complexes and supershift complexes are indicated by the arrow. C, Western blot analysis of affinity purified DNA-binding proteins. 450 mM KCl, eluted from an affinity column that contained purified protein complexes, was subjected to 10% SDS-polyacrylamide gel, transferred onto nitrocellulose membrane, and then probed with either p50 or p65 antibody.
Fig. 3. MOWSE scores from MASCOT data base searches
A, results from band 1 identifying nucleophosmin. B, results from band 2 identifying ku antigen. C, results from band 3 identifying nucleolin.
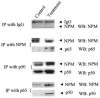
Fig. 4. Interaction of NF-_α_B proteins and NPM
In vivo binding of p50 with NPM, p65 with NPM, and NPM with p65 was confirmed by co-immunoprecipitation analysis. Combination treatment of PMA and cytokines in HepG2 cells was observed to enhance the binding of NPM with p50 and p65.
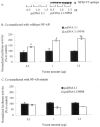
Fig. 5. NPM overexpression enhances MnSOD gene transcription
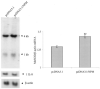
HepG2 cells were co-transfected with either NPM expression vector (pcDNA3.1/NPM) or empty vector (pcDNA3.1) along with Mn-SOD promoter- and enhancer-driven luciferase reporter vector. Parallel transfection experiments were performed using MnSOD promoter- and enhancer-driven luciferase reporter vectors, where the NF-_κ_B binding site in the enhancer region is mutated. After 12 h of co-transfection, cells were washed and grown for 24 h, and then cell lysates were collected. A, equal amounts of cellular proteins were subjected to 10% SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membrane, and then Western blot analyses were conducted with an NPM-V5-epitope antibody. B and C, luciferase activity of cell lysates was measured as a determinant of MnSOD gene transcription. Each data point represents the mean of three independent experiments ± S.D. Significantly different from corresponding empty vector transfected control; **, p < 0.01. IP, immunoprecipitation; WB, Western blot.
Fig. 6. NPM overexpression enhances MnSOD mRNA expression
HepG2 cells were transfected with 0.5 _μ_g of NPM expression vector (pcDNA3.1/NPM) or empty vector (pcDNA3.1) for 24 h. Cells were then washed with 1x PBS, and total RNA was isolated by the TRIzol method. Thirty _μ_g of RNA was subjected to 1.1% agarose gel electrophoresis and transferred onto nylon membrane. Northern analysis of RNA isolated from control and overexpressing cells were performed using a human MnSOD cDNA probe. The 1- and 4-kb human MnSOD transcripts are indicated by arrows. The same membrane was stripped and re-probed with a human IL-8 cDNA probe (middle arrow) and a _β_-actin cDNA probe (bottom arrow). The 4-kb MnSOD transcript bands were densitometrically scanned and normalized with _β_-actin. Statistical analyses were performed using three sets of experiments. Each column represents the mean of three samples ± S.D. **, p < 0.01 as compared with empty vector-transfected control.
Fig. 7. Antisense NPM overexpression suppresses MnSOD gene induction
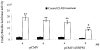
HepG2 cells were transfected with antisense NPM (pCMV/ASNPM) or empty vector (pCMV) along with MnSOD promoter-and enhancer-driven luciferase reporter vectors. After 12 h of co-transfection, cells were washed, grown for 24 h, and then treated with PMA and cytokines for 12 h, after which cells were again washed. Cell lysates were collected, and the luciferase activity was measured as a determinant of MnSOD gene transcription. Each data point represents the mean of three independent experiments ± S.D. Significant difference from corresponding control: **, p < 0.01. Significant difference from corresponding treatment group: #, p < 0.01.
Fig. 8. Antisense NPM suppresses endogenous MnSOD
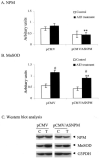
HepG2 cells were transfected with antisense NPM vector or empty vector for 12 h. Cells were washed, grown for 24 h, and then treated with PMA and cytokines for 12 h. Cells were washed, and total cell lysates were collected. Equal amounts of proteins (50 _μ_g) were subjected to 12.5% SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membrane, and then probed with MnSOD antibody. The same membranes were then stripped and re-probed with NPM (B23) antibody or GAPDH antibody, with GAPDH used as the loading control. Quantification of immunoband intensities was determined by densitometric scanning. The band intensity values of NPM and MnSOD were normalized with respect to the band intensity values of GAPDH (G3PDH). The normalized protein levels of NPM and MnSOD are shown in A and B, respectively. A representative Western blot is shown in C. Statistical analyses were performed using three sets of independent experiments. **, p < 0.01 compared with corresponding treatment; #, p < 0.01 compared with corresponding control.
Fig. 9. NPM siRNA inhibits MnSOD gene expression
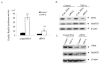
HepG2 cells were transfected with control RNA or siRNA (double-stranded siRNA in Ready pSIREN-Shuttle vector) along with MnSOD promoter-and enhancer-driven luciferase reporter vectors for 24 h. Cells were then washed with 1x PBS and incubated in medium at 37 °C. After 12 h of co-transfection, cells were treated with TNF-α (200 units/ml) for 12 h, and then cell lysates were collected and luciferase activity was measured. A, control RNA or NPM siRNA transfected into HepG2 cells was used to isolate total RNA or total cell lysates. B, RT-PCR of RNA isolated from control and TNF-α (200 units/ml for 12 h) -treated cells was carried out using primers for each specific gene as described under “Experimental Procedures.” C, proteins from total cellular extracts were analyzed by 10% SDS-PAGE, transferred onto nitrocellulose membrane, and probed with MnSOD antibody. The same membrane was re-probed with NPM antibody (B23) or _β_-actin antibody. Statistical analyses were performed in three independent experiments. **, p < 0.01 compared with the corresponding control; #, p < 0.01 compared with the corresponding treatment.
Fig. 10. The role of NPM on mRNA levels of NF-_α_B-targeted genes
A, schematics of transcription factor binding sites in the human MnSOD and IL-8 gene promoters. Corresponding transcription factor binding sites for key transcription factors in the 5′-flanking and enhancer regions are indicated by vertical lines. In the promoter region of the IL-8 gene, typical TATA and CAAT signal sequences are highlighted. B, RT-PCR products of RNA isolated from NPM siRNA transfected control and treated cells as described in the legend to Fig. 9.
Similar articles
- Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2.
Kiningham KK, Xu Y, Daosukho C, Popova B, St Clair DK. Kiningham KK, et al. Biochem J. 2001 Jan 1;353(Pt 1):147-156. Biochem J. 2001. PMID: 11115408 Free PMC article. - Specificity protein 1-dependent p53-mediated suppression of human manganese superoxide dismutase gene expression.
Dhar SK, Xu Y, Chen Y, St Clair DK. Dhar SK, et al. J Biol Chem. 2006 Aug 4;281(31):21698-21709. doi: 10.1074/jbc.M601083200. Epub 2006 Jun 1. J Biol Chem. 2006. PMID: 16740634 Free PMC article. - IkappaBalpha (inhibitory kappaBalpha) identified as labile repressor of MnSOD (manganese superoxide dismutase) expression.
Kiningham KK, Daosukho C, St Clair DK. Kiningham KK, et al. Biochem J. 2004 Dec 15;384(Pt 3):543-9. doi: 10.1042/BJ20040714. Biochem J. 2004. PMID: 15330761 Free PMC article. - Chronic exposure to 12-O-tetradecanoylphorbol-13-acetate represses sod2 induction in vivo: the negative role of p50.
Dhar SK, Xu Y, Noel T, St Clair DK. Dhar SK, et al. Carcinogenesis. 2007 Dec;28(12):2605-13. doi: 10.1093/carcin/bgm163. Epub 2007 Jul 25. Carcinogenesis. 2007. PMID: 17652337 Free PMC article. - The NF-κB Nucleolar Stress Response Pathway.
Thoms HC, Stark LA. Thoms HC, et al. Biomedicines. 2021 Aug 25;9(9):1082. doi: 10.3390/biomedicines9091082. Biomedicines. 2021. PMID: 34572268 Free PMC article. Review.
Cited by
- Identification of proteins differentially expressed in adriamycin-resistant (pumc-91/ADM) and parental (pumc-91) human bladder cancer cell lines by proteome analysis.
Meng Q, Lei T, Zhang M, Zhao J, Zhao XH, Zhang M. Meng Q, et al. J Cancer Res Clin Oncol. 2013 Mar;139(3):509-19. doi: 10.1007/s00432-012-1350-8. Epub 2012 Nov 27. J Cancer Res Clin Oncol. 2013. PMID: 23183654 - Nucleophosmin acts as a novel AP2alpha-binding transcriptional corepressor during cell differentiation.
Liu H, Tan BC, Tseng KH, Chuang CP, Yeh CW, Chen KD, Lee SC, Yung BY. Liu H, et al. EMBO Rep. 2007 Apr;8(4):394-400. doi: 10.1038/sj.embor.7400909. Epub 2007 Feb 23. EMBO Rep. 2007. PMID: 17318229 Free PMC article. - Fine-tuning nucleophosmin in macrophage differentiation and activation.
Guery L, Benikhlef N, Gautier T, Paul C, Jego G, Dufour E, Jacquel A, Cally R, Manoury B, Vanden Berghe T, Vandenabeele P, Droin N, Solary E. Guery L, et al. Blood. 2011 Oct 27;118(17):4694-704. doi: 10.1182/blood-2011-03-341255. Epub 2011 Aug 29. Blood. 2011. PMID: 21876121 Free PMC article. - PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation.
Gao F, Kwon SW, Zhao Y, Jin Y. Gao F, et al. J Biol Chem. 2009 Aug 14;284(33):22263-22273. doi: 10.1074/jbc.M109.033118. Epub 2009 Jun 16. J Biol Chem. 2009. PMID: 19531481 Free PMC article.
References
- Halliwel B, Gutteridge JM. Methods Enzymol. 1990;186:1–85. - PubMed
- Dougall WC, Nick HS. Endocrinology. 1991;129:2379–2384. - PubMed
- Suzuki K, Tatsumi H, Satoh S, Sendra T, Naskata T, Fuji J, Takiguchi N. Am. J. Physiol. 1993;265:H1173–H1178. - PubMed
- Del Maestro R, McDonald W. Mech. Aging Dev. 1989;48:15–31. - PubMed
- Wong GH, Elwell J, Overly L, Goeddel DV. Cell. 1989;58:923–931. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HL 03544/HL/NHLBI NIH HHS/United States
- P01 AG005119/AG/NIA NIH HHS/United States
- CA 59835/CA/NCI NIH HHS/United States
- R01 CA049797-13/CA/NCI NIH HHS/United States
- CA 49797/CA/NCI NIH HHS/United States
- R01 CA049797/CA/NCI NIH HHS/United States
- AG 05119/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases