Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT - PubMed (original) (raw)
Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT
Yanping Wang et al. J Bacteriol. 2004 May.
Abstract
CTnDOT is a conjugative transposon (CTn) that is found in many Bacteroides strains. Transfer of CTnDOT is stimulated 100- to 1,000-fold if the cells are first exposed to tetracycline (TET). Both excision and transfer of CTnDOT are stimulated by TET. An operon that contains a TET resistance gene, tetQ, and two regulatory genes, rteA and rteB, is essential for control of excision and transfer functions. At first, it appeared that RteA and RteB, which are members of a two-component regulatory system, might be directly responsible for the TET effect. We show here, however, that neither RteA nor RteB affected expression of the operon. TetQ, a ribosome protection type of TET resistance protein, actually reduced operon expression, possibly by interacting with ribosomes that are translating the tetQ message. Fusions of tetQ with a reporter gene, uidA, were only expressed at a high level when the fusion was cloned in frame with the first six codons of tetQ. However, out of frame fusions or fusions ending at the other five codons of tetQ showed much lower expression of the uidA gene. Moreover, reverse transcription-PCR amplification of tetQ mRNA revealed that despite the fact that the uidA gene product, beta-glucuronidase (GUS), was produced only when the cells were exposed to TET, tetQ mRNA was produced in both the presence and absence of TET. Computer analysis of the region upstream of the tetQ start codon predicted that the mRNA in this region could form a complex RNA hairpin structure that would prevent access of ribosomes to the ribosome binding site. Mutations that abolished base pairing in the stem that formed the base of this putative hairpin structure made GUS production as high in the absence of TET as in TET-stimulated cells. Compensatory mutations that restored the hairpin structure led to a return of regulated production of GUS. Thus, the tetQ-rteA-rteB operon appears to be regulated by a translational attenuation mechanism.
Figures
FIG. 1.
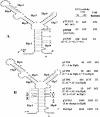
(A) Organization of CTnDOT. The names of the genes are given above the arrows. The arrows indicate the orientations and positions of genes on CTnDOT. The size of each functional region is given under the diagram. Note, the size is not in ratio. (B) Diagram of the central regulatory region that includes the tetQ rteA rteB operon and the rteC gene and description of their functions. Pq and Pc are the tetQ promoter and the rteC promoter, respectively. Numbers 1, 2, and 3 indicate three levels of regulation. Tc, TET.
FIG. 2.
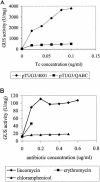
GUS activities of different Pq-uidA fusions in strain BT4004 (which contains CTnERL). The increase is the GUS activity detected in TET-induced cells (+Tc) divided by the GUS activity in noninduced cells (−Tc). The thick arrows indicate the reporter gene uidA. The hollow arrows in pTUG10, pTUG1, and pTUG12 indicate an out of frame fusion to the uidA gene. The gray arrows in pTUG3 and pTUG11 indicate an in-frame fusion to the uidA gene. The numbers at the left corner indicate positions relative to the start codon of tetQ. TET (1 μg/ml) was used to induce expression. S, SstI; E, EcoNI; B, BalI; R, EcoRI.
FIG. 3.
Determination of which CTnDOT genes affect TET (Tc)-induced expression through the Pq promoter. pTUG11 and pTUG3 were used to determine GUS production in different backgrounds. The strain labeled “QABC in chromosome” (BT4001ΩQABC) had a single copy of QABC integrated into the chromosome of BT4001. The strain labeled “QABC in plasmid” (pAMS9) had 5 to 10 copies of QABC per cell. The strain labeled “Q only in plasmid” (pTC-COW) contained only the tetQ gene on a plasmid (5 to 10 copies per cell). The strain labeled “Pq only in plasmid” (pYP44) had only the Pq region on a plasmid (5 to 10 copies per cell). MIC indicates the minimal inhibition concentration of TET. As in Fig. 2, the increase is the GUS activity detected in TET-induced cells (+Tc) divided by the GUS activity in noninduced cells (−Tc).
FIG. 4.
GUS production under different conditions. (A) GUS activity of pTUG3/4001 with or without the tetQ gene in the strain. The GUS activity associated with pTUG3 without the tetQ gene was much higher than that associated with tetQ. The MIC for pTUG3/4001 (Genr Emr) is 0.12 μg/ml. Thus, the induction concentration of TET (Tc) ranges from 0 to 0.10 μg/ml. (B) GUS activity of pYP67/4001 (Genr Cefr) induced by LIN (0 to 0.6 μg/ml), ERY (0 to 0.1 μg/ml), and CHL (0 to 0.5 μg/ml). pYP67 contains the Pq-uidA fusion in a vector that carries a FOX resistance gene.
FIG. 5.
(A) Localization of Pq promoter. The number before each fusion indicates the relative position of the fusion joint to the ATG start codon of tetQ. The name of each fusion is labeled behind the number. The consensus tetQ promoter sequence identified by Bayley et al. is indicated by (−33, −7). The thick line in pYP64 indicates that there are four point mutations (Fig. 5B) within the −7 region of the Pq promoter that eliminate the consensus −7 sequence. Tc, TET. (B) Nucleotide sequence of the 209 bp upstream of the ATG start codon of tetQ. The asterisk above the sequence indicates the position of the TIS. The Pq region is indicated by a −33 region and a −7 region. Arrows labeled Hp1 and Hp8 indicate that a hairpin can be formed in the mRNA by Hp1 and Hp8. The numbers above the vertical lines indicate their positions relative to the ATG start codon of tetQ. The deduced amino acid sequences of two leader peptides are given in single-letter code under the nucleotide sequence. The bold ATG is the putative start codon of tetQ. Asterisks indicate stop codons.
FIG. 6.
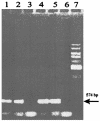
RT-PCR to detect the amount of tetQ mRNA in BT4107 under TET-induced and non-TET-induced conditions. Lanes 1 and 2, tetQ product amplified by RT-PCR within the no-TET cells; lane 3, control to which RT was not added; lanes 4 and 5, tetQ product amplified by RT-PCR from the TET-treated BT4107 cells; lane 6, control to which RT was not added; lane 7, 1-kb standard. The arrow indicates the size of the amplified tetQ product (574 bp).
FIG. 7.
Determination of the start codon of the tetQ gene. The Pq regions in all five of the constructs start from −333 bp upstream of tetQ, but each ends at a different position. The Pq region in pTUG13-in3 ends at the GTG putative start codon of tetQ. In pTUG13-in2, the ATG start codon was fused to the uidA gene. In both pTUG13-in1 and pYP48, the Pq region ends at the sixth amino acid of TetQ and was fused to the uidA gene. The Pq region in pYP48 is the same as that in pTUG13-in1 except that two stop codons, which are indicated by a circle, were put between GTG and ATG, their exact positions are shown in Fig. 8A. Tc, TET.
FIG. 8.
Evidence for the importance of a secondary structure of the 130-bp tetQ mRNA in TET (Tc) induction. Asterisks indicate the TIS; Hp1 to Hp8 indicate that their mRNA sequences can be part of a hairpin. The Δ_G_ of the secondary structure of the wild-type Pq mRNA (−130 to +4) is −33.24 kcal/mol. The nucleotides following the dotted lines indicate substituted nucleotides to disrupt or reestablish the secondary structure. (A) GUS activity of the wild-type strain and mutants, which have a partially disrupted hairpin in the Pq region. (B) Evidence to show that the Hp1- and Hp8-formed hairpin is responsible for all of the TET induction activity. Strain Del Hp1 is also named pYP220. nt, nucleotide.
Similar articles
- Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT.
Wang Y, Rotman ER, Shoemaker NB, Salyers AA. Wang Y, et al. J Bacteriol. 2005 Apr;187(8):2673-80. doi: 10.1128/JB.187.8.2673-2680.2005. J Bacteriol. 2005. PMID: 15805513 Free PMC article. - Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT.
Moon K, Shoemaker NB, Gardner JF, Salyers AA. Moon K, et al. J Bacteriol. 2005 Aug;187(16):5732-41. doi: 10.1128/JB.187.16.5732-5741.2005. J Bacteriol. 2005. PMID: 16077120 Free PMC article. - An unexpected effect of tetracycline concentration: growth phase-associated excision of the Bacteroides mobilizable transposon NBU1.
Song B, Wang GR, Shoemaker NB, Salyers AA. Song B, et al. J Bacteriol. 2009 Feb;191(3):1078-82. doi: 10.1128/JB.00637-08. Epub 2008 Oct 24. J Bacteriol. 2009. PMID: 18952794 Free PMC article. - Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events.
Waters JL, Salyers AA. Waters JL, et al. mBio. 2013 Oct 29;4(6):e00569-13. doi: 10.1128/mBio.00569-13. mBio. 2013. PMID: 24169574 Free PMC article. Review. - Chromosomal gene transfer elements of the Bacteroides group.
Salyers AA, Shoemaker NB. Salyers AA, et al. Eur J Clin Microbiol Infect Dis. 1992 Nov;11(11):1032-8. doi: 10.1007/BF01967795. Eur J Clin Microbiol Infect Dis. 1992. PMID: 1338314 Review.
Cited by
- Tetracycline-related transcriptional regulation of the CTnDOT mobilization region.
Waters JL, Wang GR, Salyers AA. Waters JL, et al. J Bacteriol. 2013 Dec;195(24):5431-8. doi: 10.1128/JB.00691-13. Epub 2013 Sep 27. J Bacteriol. 2013. PMID: 24078614 Free PMC article. - Comprehensive analyses of a large human gut Bacteroidales culture collection reveal species- and strain-level diversity and evolution.
Zhang ZJ, Cole CG, Coyne MJ, Lin H, Dylla N, Smith RC, Pappas TE, Townson SA, Laliwala N, Waligurski E, Ramaswamy R, Woodson C, Burgo V, Little JC, Moran D, Rose A, McMillin M, McSpadden E, Sundararajan A, Sidebottom AM, Pamer EG, Comstock LE. Zhang ZJ, et al. Cell Host Microbe. 2024 Oct 9;32(10):1853-1867.e5. doi: 10.1016/j.chom.2024.08.016. Epub 2024 Sep 17. Cell Host Microbe. 2024. PMID: 39293438 - Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT.
Jeters RT, Wang GR, Moon K, Shoemaker NB, Salyers AA. Jeters RT, et al. J Bacteriol. 2009 Oct;191(20):6374-82. doi: 10.1128/JB.00739-09. Epub 2009 Aug 21. J Bacteriol. 2009. PMID: 19700528 Free PMC article. - Translational control of tetracycline resistance and conjugation in the Bacteroides conjugative transposon CTnDOT.
Wang Y, Rotman ER, Shoemaker NB, Salyers AA. Wang Y, et al. J Bacteriol. 2005 Apr;187(8):2673-80. doi: 10.1128/JB.187.8.2673-2680.2005. J Bacteriol. 2005. PMID: 15805513 Free PMC article. - First Report of Integrative Conjugative Elements in Riemerella anatipestifer Isolates From Ducks in China.
Zhu D, Wan J, Yang Z, Xu J, Wang M, Jia R, Chen S, Liu M, Zhao X, Yang Q, Wu Y, Zhang S, Liu Y, Zhang L, Yu Y, Chen X, Cheng A. Zhu D, et al. Front Vet Sci. 2019 Apr 24;6:128. doi: 10.3389/fvets.2019.00128. eCollection 2019. Front Vet Sci. 2019. PMID: 31069241 Free PMC article.
References
- Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. - PubMed
- Burdett, V. 1991. Purification and characterization of Tet(M), a protein that renders ribosomes resistant to tetracycline. J. Biol. Chem. 266:2872-2877. - PubMed
- Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for the excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources