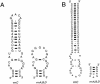Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons - PubMed (original) (raw)
Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons
Silvia G Acinas et al. J Bacteriol. 2004 May.
Abstract
The level of sequence heterogeneity among rrn operons within genomes determines the accuracy of diversity estimation by 16S rRNA-based methods. Furthermore, the occurrence of widespread horizontal gene transfer (HGT) between distantly related rrn operons casts doubt on reconstructions of phylogenetic relationships. For this study, patterns of distribution of rrn copy numbers, interoperonic divergence, and redundancy of 16S rRNA sequences were evaluated. Bacterial genomes display up to 15 operons and operon numbers up to 7 are commonly found, but approximately 40% of the organisms analyzed have either one or two operons. Among the Archaea, a single operon appears to dominate and the highest number of operons is five. About 40% of sequences among 380 operons in 76 bacterial genomes with multiple operons were identical to at least one other 16S rRNA sequence in the same genome, and in 38% of the genomes all 16S rRNAs were invariant. For Archaea, the number of identical operons was only 25%, but only five genomes with 21 operons are currently available. These considerations suggest an upper bound of roughly threefold overestimation of bacterial diversity resulting from cloning and sequencing of 16S rRNA genes from the environment; however, the inclusion of genomes with a single rrn operon may lower this correction factor to approximately 2.5. Divergence among operons appears to be small overall for both Bacteria and Archaea, with the vast majority of 16S rRNA sequences showing <1% nucleotide differences. Only five genomes with operons with a higher level of nucleotide divergence were detected, and Thermoanaerobacter tengcongensis exhibited the highest level of divergence (11.6%) noted to date. Overall, four of the five extreme cases of operon differences occurred among thermophilic bacteria, suggesting a much higher incidence of HGT in these bacteria than in other groups.
Figures
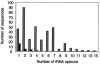
FIG. 1.
Distribution of different rrn operon numbers among bacterial (gray bars) and archaeal (black bars) isolates. Data were obtained from complete genome sequences from the NCBI and TIGR genome databases, the rrndb database, and the literature.
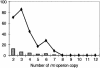
FIG. 2.
Numbers (bars) and percentages of the total (line) of genomes with all identical 16S rRNA sequences among operons. Data were retrieved from the NCBI and TIGR genome databases and the rrndb database.
FIG. 3.
Secondary structure model of the helices that differ in length between operons in Thermoanaerobacter tengcongensis. Nucleotide positions 444 to 490 (A) and 1,447 to 1,456 (B) are given according to Escherichia coli reference numbering.
References
- Alm, R. A., L.-S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human astric pathogen Helicobacter pylori. Nature 397:176-180. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases