A new set of BXD recombinant inbred lines from advanced intercross populations in mice - PubMed (original) (raw)
A new set of BXD recombinant inbred lines from advanced intercross populations in mice
Jeremy L Peirce et al. BMC Genet. 2004.
Abstract
Background: Recombinant inbred (RI) strains are an important resource for mapping complex traits in many species. While large RI panels are available for Arabidopsis, maize, C. elegans, and Drosophila, mouse RI panels typically consist of fewer than 30 lines. This is a severe constraint on the power and precision of mapping efforts and greatly hampers analysis of epistatic interactions.
Results: In order to address these limitations and to provide the community with a more effective collaborative RI mapping panel we generated new BXD RI strains from two independent advanced intercrosses (AI) between C57BL/6J (B6) and DBA/2J (D2) progenitor strains. Progeny were intercrossed for 9 to 14 generations before initiating inbreeding, which is still ongoing for some strains. Since this AI base population is highly recombinant, the 46 advanced recombinant inbred (ARI) strains incorporate approximately twice as many recombinations as standard RI strains, a fraction of which are inevitably shared by descent. When combined with the existing BXD RI strains, the merged BXD strain set triples the number of previously available unique recombinations and quadruples the total number of recombinations in the BXD background.
Conclusion: The combined BXD strain set is the largest mouse RI mapping panel. It is a powerful tool for collaborative analysis of quantitative traits and gene function that will be especially useful to study variation in transcriptome and proteome data sets under multiple environments. Additional strains also extend the value of the extensive phenotypic characterization of the previously available strains. A final advantage of expanding the BXD strain set is that both progenitors have been sequenced, and approximately 1.8 million SNPs have been characterized. This provides unprecedented power in screening candidate genes and can reduce the effective length of QTL intervals. It also makes it possible to reverse standard mapping strategies and to explore downstream effects of known sequence variants.
Figures
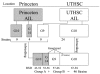
Figure 1
ARI breeding schematic. This figure shows the breeding history, genetic derivations, and genotyping status of the BXD ARI strain set. We commonly refer to animals derived from the BXD AIL generated at Princeton University as "Group A" and animals derived from the BXD AIL generated at the University of Tennessee Health Science Center as "Group B." Group A animals are shown in gray. Genotyping refers to our dense, 588 marker effort. Nearly all strains in both groups are genotyped at 268 loci across the genome.
Figure 2
ARI chromosome 15. Chromosome 15 in 22 genotyped ARI strains and the 34 BXD RI strains. Dark grey regions are homozygous B6, white regionsare homozygous DBA, and light grey regions are heterozygous. All BXD data are taken from [4] and are publicly accessible via
. Number of recombinations per strain is displayed below each strain. Where heterozygous regions were present, the number of recombinations was calculated based on the smallest number of recombinations that might ultimately be resolved from that pattern. For example BXD47 has 0 recombinations rather than 2. Even discounting additional recombinations from resolution of heterozygous regions, the ARI animals have a considerably larger number of recombinations, 3.0 per strain compared to 1.3 per strain in BXD animals for Chr 15. BXD37, BXD41, BXD47, and BXD58, are now extinct.
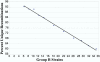
Figure 3
Population size and unique recombinations in the UTHSC ARI lines. Fraction of unique recombinations as a function of number of strains wassampled in the set of UTHSC-derived strains. We derived each data point in this highly conservative estimate by randomly sampling the indicated number of strains from the UTHSC-derived strain set, using 100 samples per data point. Up to one directional transition from B6 to DBA and the reverse was scored for each marker interval and the total was divided by the number of intervals with more than 0 recombinations. Heterozygous regions were handled as described for Fig. 2. Fraction of unique recombinations depends on a number of variables including AIL population size, so other crosses may differ.
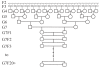
Figure 4
New RI-like breeding scheme. Novel proposed method for maximizing unique recombinations archived in a 2-way RI-like cross. The breeding scheme shown above and discussed more generally in the text results in a single strain with 75% more unique recombinations than a standard RI strain. This example begins with 7 generations of crosses (parental intercross and F1 × F1 cross not shown) prior to inbreeding. All recombinations generated using this method are independent.
Similar articles
- Using gene expression databases for classical trait QTL candidate gene discovery in the BXD recombinant inbred genetic reference population: mouse forebrain weight.
Lu L, Wei L, Peirce JL, Wang X, Zhou J, Homayouni R, Williams RW, Airey DC. Lu L, et al. BMC Genomics. 2008 Sep 25;9:444. doi: 10.1186/1471-2164-9-444. BMC Genomics. 2008. PMID: 18817551 Free PMC article. - Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps.
Taylor BA, Wnek C, Kotlus BS, Roemer N, MacTaggart T, Phillips SJ. Taylor BA, et al. Mamm Genome. 1999 Apr;10(4):335-48. doi: 10.1007/s003359900998. Mamm Genome. 1999. PMID: 10087289 - Quantitative trait loci affecting ethanol sensitivity in BXD recombinant inbred mice.
Browman KE, Crabbe JC. Browman KE, et al. Alcohol Clin Exp Res. 2000 Jan;24(1):17-23. Alcohol Clin Exp Res. 2000. PMID: 10656187 - BXD Recombinant Inbred Mice as a Model to Study Neurotoxicity.
Martins AC, López-Granero C, Ferrer B, Tinkov AA, Skalny AV, Paoliello MMB, Aschner M. Martins AC, et al. Biomolecules. 2021 Nov 25;11(12):1762. doi: 10.3390/biom11121762. Biomolecules. 2021. PMID: 34944406 Free PMC article. Review. - Use of recombinant inbred strains for studying genetic determinants of responses to alcohol.
Crabbe JC, Belknap JK, Buck KJ, Metten P. Crabbe JC, et al. Alcohol Alcohol Suppl. 1994;2:67-71. Alcohol Alcohol Suppl. 1994. PMID: 8974318 Review.
Cited by
- A novel mechanism regulating insulin secretion involving Herpud1 in mice.
Wong N, Morahan G, Stathopoulos M, Proietto J, Andrikopoulos S. Wong N, et al. Diabetologia. 2013 Jul;56(7):1569-76. doi: 10.1007/s00125-013-2908-y. Epub 2013 Apr 26. Diabetologia. 2013. PMID: 23620059 - Systems genetics identifies Hp1bp3 as a novel modulator of cognitive aging.
Neuner SM, Garfinkel BP, Wilmott LA, Ignatowska-Jankowska BM, Citri A, Orly J, Lu L, Overall RW, Mulligan MK, Kempermann G, Williams RW, O'Connell KM, Kaczorowski CC. Neuner SM, et al. Neurobiol Aging. 2016 Oct;46:58-67. doi: 10.1016/j.neurobiolaging.2016.06.008. Epub 2016 Jun 17. Neurobiol Aging. 2016. PMID: 27460150 Free PMC article. - Expression QTL mapping in regulatory and helper T cells from the BXD family of strains reveals novel cell-specific genes, gene-gene interactions and candidate genes for auto-immune disease.
Alberts R, Chen H, Pommerenke C, Smit AB, Spijker S, Williams RW, Geffers R, Bruder D, Schughart K. Alberts R, et al. BMC Genomics. 2011 Dec 19;12:610. doi: 10.1186/1471-2164-12-610. BMC Genomics. 2011. PMID: 22182475 Free PMC article. - Bone morphology in 46 BXD recombinant inbred strains and femur-tibia correlation.
Zhang Y, Huang J, Jiao Y, David V, Kocak M, Roan E, Di'Angelo D, Lu L, Hasty KA, Gu W. Zhang Y, et al. ScientificWorldJournal. 2015;2015:728278. doi: 10.1155/2015/728278. Epub 2015 Feb 25. ScientificWorldJournal. 2015. PMID: 25811045 Free PMC article. - Genome-level analysis of genetic regulation of liver gene expression networks.
Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, Threadgill DW, Rusyn I. Gatti D, et al. Hepatology. 2007 Aug;46(2):548-57. doi: 10.1002/hep.21682. Hepatology. 2007. PMID: 17542012 Free PMC article.
References
- Taylor BA. Recombinant inbred strains. In: Lyon M L and Searle A G, editor. Genetic Variation in the Laboratory Mouse, 2nd edn. Oxford, Oxford University Press; 1989. pp. 773–796.
- Silver LM. Mouse Genetics: Concepts and Applications. New York, Oxford University Press; 1995.
Publication types
MeSH terms
Grants and funding
- P20 MH062009/MH/NIMH NIH HHS/United States
- R37 HD020275/HD/NICHD NIH HHS/United States
- R37 HD20275/HD/NICHD NIH HHS/United States
- U01 AA013499/AA/NIAAA NIH HHS/United States
- U01AA13499/AA/NIAAA NIH HHS/United States
- U24AA135B/AA/NIAAA NIH HHS/United States
- P20-MH62009/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases