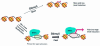The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming - PubMed (original) (raw)
The chromatin-remodeling BAF complex mediates cellular antiviral activities by promoter priming
Kairong Cui et al. Mol Cell Biol. 2004 May.
Abstract
The elicitation of cellular antiviral activities is dependent on the rapid transcriptional activation of interferon (IFN) target genes. It is not clear how the interferon target promoters, which are organized into chromatin structures in cells, rapidly respond to interferon or viral stimulation. In this report, we show that alpha IFN (IFN-alpha) treatment of HeLa cells induced hundreds of genes. The induction of the majority of these genes was inhibited when one critical subunit of the chromatin-remodeling SWI/SNF-like BAF complexes, BAF47, was knocked down via RNA interference. Inhibition of BAF47 blocked the cellular response to viral infection and impaired cellular antiviral activity by inhibiting many IFN- and virus-inducible genes. We show that the BAF complex was required to mediate both the basal-level expression and the rapid induction of the antiviral genes. Further analyses indicated that the BAF complex primed some IFN target promoters by utilizing ATP-derived energy to maintain the chromatin in a constitutively open conformation, allowing faster and more potent induction after IFN-alpha treatment. We propose that constitutive binding of the BAF complex is an important mechanism for the IFN-inducible promoters to respond rapidly to IFN and virus stimulation.
Figures
FIG. 1.
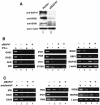
The BAF complex is required for cellular responses to IFN-α and poly(I)/poly(C) stimulation. (A) BAF47 was efficiently knocked down by RNA interference. HeLa cells were transfected with a control vector (lane 1) or a siRNA construct targeting BAF47 (lane 2) and selected for 2 days in puromycin. The remaining cells were lysed and analyzed by Western blotting with antibodies against BAF47, BRG1, BRM, or β actin. (B) The BAF complex is required for the induction of IFN target genes by IFN-α. HeLa cells were transfected with the siRNA construct targeting BAF47 (siBAF47) and selected for 2 days with puromycin. Following stimulation with 500 U of IFN-α/ml for 8 h, the total RNAs were isolated for RT-PCR analysis with the primers indicated on the left sides of the panels. (C) The BAF complex is required for the induction of IFN target genes by poly(I)/poly(C). HeLa cells were transfected and selected as described above. Following stimulation with poly(I)/poly(C), total RNAs were isolated and analyzed as described above.
FIG. 2.
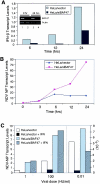
The BAF complex is required for the induction of cellular antiviral activities. (A) Expression of IFN-β upon NDV infection. Control or BAF-inhibited (siBAF47) HeLa cells were infected with NDV (100 HU/ml) for 1 h, and IFN-β levels at the indicated time points were measured by real-time PCR. The inset shows the results of RT-PCR analyses of BAF47 and β actin mRNAs after 0 and 24 h of viral infection in HeLa cells transfected with the control vector (lanes 1 and 3) or the siBAF47 construct (lanes 2 and 4). (B) Assessment of NDV replication. Cells were infected with NDV, and NDV NP transcript levels were measured at the indicated time points by real-time PCR. The increase (fold) is shown. (C) Effect of IFN-α on NDV replication. Cells were infected with different doses of NDV (0.01, 1, and 100 HU/ml) in the presence or absence of IFN-α (1,000 U/ml), and viral replication by NDV NP was assessed by real-time PCR at 12 h after infection. Values on the right y axis correspond to transcript levels obtained with 0.01 HU/ml, while those on the left y axis correspond to transcript levels with 1 and 100 HU of NDV/ml.
FIG. 3.
The BAF complex regulates IFITM1 promoter activity by modulating its chromatin structure. (A) Inhibiting the BAF complexes abolished the induction of the IFITM1 promoter by IFN-α. HeLa cells were transfected with the IFITM1 promoter reporter construct, pREP4-IFITM1pr-luc, together with a control vector or the siRNA construct targeting BAF47 (siBAF47) for 48 to 72 h. Following treatment with 500 U of IFN-α/ml for 12 h, the luciferase activity was analyzed with a dual luciferase system from Promega. (B) Knockdown of BAF47 did not inhibit claudin promoter activity. HeLa cells were transfected with the claudin promoter reporter construct and analyzed as described for panel A. (C) The IFITM1 promoter has a positioned nucleosome. The genomic DNA or nuclei isolated from HeLa or SW-13 cells was digested with microccocal nuclease. The double-stranded cleavages in the nucleosomal linker regions were detected by ligation-mediated PCR with primers specific for the IFITM1 promoter sequence (PCR primer −273F [5′ACAGTGAGGTCCTGTACTTGCTGG3′] and labeling primer −261F [5′TGTACTTGCTGGCCTGGGGTG3′]). The open oval on the right indicates the regions protected by the nucleosomal structure. The transcription start site (+1) and direction are indicated. The filled square indicates a potential binding site for ISGF3 (ISRE). The recognition sites for the restriction enzymes (AvaII and HgiAI) are indicated. (D) The BAF complex is required for the chromatin remodeling of the IFITM1 promoter upon IFN-α stimulation. HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. After stimulation with 500 U of IFN-α/ml for 2 h, the cells were permeabilized and briefly digested with AvaII enzyme, followed by complete digestion of the purified genomic DNA with HgiAI enzyme. The cleavage sites were detected by linker ligation-mediated PCR with IFITM1 promoter-specific primers. The data were quantified by PhophorImager analysis. The intensity of the bands produced by AvaII digestion after normalization to the HgiAI digestion is indicated below the panel. The experiments were repeated three times, and similar results were obtained. Size markers are indicated on the left side of the panel. (E) Knocking down BAF47 reduced histone H4 acetylation at the IFITM1 promoter. The HeLa cells were transfected with control vector or siRNA targeting BAF47 (siBAF47) and selected as described in the legend to Fig. 1. The chromatin lysates were prepared and immunoprecipitated with preimmune serum and antibodies against tetra-acetylated histone H4 tail as described previously (24). The IFITM1 promoter sequence and the CSF1 upstream sequence (control) in the immunoprecipitated DNA were analyzed by PCR. The chromatin (chr) input was diluted three times at each step.
FIG. 4.
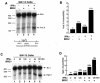
Expression of BRG1 results in more rapid kinetics and higher levels of the IFITM1 gene induction in SW-13 cells in response to IFN-α. (A) Analysis of IFITM1 mRNA expression induced by treatment with IFN-α. Total RNAs extracted from HeLa cells and SW-13 cells treated with IFN-α were reverse transcribed, amplified by PCR with IFITM1 primers, slot blotted onto nylon membrane, and detected by hybridization with a 32P-labeled IFITM1 cDNA probe. (B) The slot blot was quantified by PhosphorImager analysis and plotted after normalization to β actin signals. (C) The same samples were amplified with ISG15 primers and analyzed as described above. (D) SW-13 cells were transfected with pBJ5 or pBJ5-BRG1 for 24 h, followed by treatment with 500 U of IFN-α/ml for various times. The total RNAs were analyzed as described for panel A. (E) Quantification of the samples in panel D by PhosphorImager analysis.
FIG. 5.
Expression of BRG1 controls both basal and induced chromatin remodeling of the IFITM1 promoter. (A) Nuclei isolated from SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h before treatment with IFN-α for 2 h were briefly digested with AvaII. The purified genomic DNA was digested to completion with BclI. The cleavage sites were detected by linker ligation-mediated PCR with the IFITM1 promoter-specific primers as described in the legend to Fig. 3B. (B) Quantification of the data in panel A by PhosphorImager analysis. (C) Nuclei isolated from SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h before treatment with IFN-α for various times were briefly digested with HgiAI and analyzed as described for panel A. (D) Quantification of the data in panel C by PhosphorImager analysis. The experiments were repeated three times, with similar results.
FIG. 6.
BRG1 is constitutively associated with the IFITM1 promoter. (A) SW-13 cells transfected with pBJ5 or pBJ5-BRG1 for 24 h were treated with 500 U of IFN-α/ml for indicated periods of time. Chromatin lysates were prepared and immunoprecipitated as described in the legend to Fig. 3C with the antibodies indicated on the left side of the panel. After reverse cross-linking, the immunoprecipitated DNA was analyzed by PCR with primers specific to the IFITM1 promoter region (Pr), the 3′ untranslated region (3′U), or the 5′ far-upstream region of the CSF1 gene (5′UCSF). The quantification of the data is shown below each panel. (B) The IFITM1 promoter in the pREP4 reporter vector was cotransfected with pBJ5 or pBJ5-BRG1 into SW-13 cells. Luciferase activity was determined after 24 h of transfection. (C) BRG1 is constitutively associated with the IFITM1 promoter in HeLa cells even without IFN-α stimulation. HeLa cells were stimulated with 500 U of IFN-α/ml for 30 min, followed by chromatin preparation and chromatin immunoprecipitation as described in the legend to Fig. 3C. The purified DNA was analyzed by PCR with the IFITM1 promoter primers (top two panels) or primers for the 3′ untranslated region of the IFITM2 gene (bottom panel). The chromatin (chr) input was diluted 5 times at each step. The preimmune serum (PreIm) and antibodies are indicated above the panels. The quantification of the data after normalization to the preimmune serum is shown below each panel.
FIG. 7.
The BAF complex primes the chromatin structure of IFN target promoters for more rapid kinetics and higher levels of induction in response to stimuli.
Similar articles
- Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex.
Liu H, Kang H, Liu R, Chen X, Zhao K. Liu H, et al. Mol Cell Biol. 2002 Sep;22(18):6471-9. doi: 10.1128/MCB.22.18.6471-6479.2002. Mol Cell Biol. 2002. PMID: 12192045 Free PMC article. - Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast.
Ansari SA, Paul E, Sommer S, Lieleg C, He Q, Daly AZ, Rode KA, Barber WT, Ellis LC, LaPorta E, Orzechowski AM, Taylor E, Reeb T, Wong J, Korber P, Morse RH. Ansari SA, et al. J Biol Chem. 2014 May 23;289(21):14981-95. doi: 10.1074/jbc.M113.529354. Epub 2014 Apr 11. J Biol Chem. 2014. PMID: 24727477 Free PMC article. - SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters.
Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, Cassel SH, Qing H, Widmer CJ, Demetri GD, Irizarry RA, Zhao K, Ranish JA, Kadoch C. Nakayama RT, et al. Nat Genet. 2017 Nov;49(11):1613-1623. doi: 10.1038/ng.3958. Epub 2017 Sep 25. Nat Genet. 2017. PMID: 28945250 Free PMC article. - BAFfling pathologies: Alterations of BAF complexes in cancer.
Arnaud O, Le Loarer F, Tirode F. Arnaud O, et al. Cancer Lett. 2018 Apr 10;419:266-279. doi: 10.1016/j.canlet.2018.01.046. Epub 2018 Jan 31. Cancer Lett. 2018. PMID: 29374542 Review. - Chromatin remodeling during glucocorticoid receptor regulated transactivation.
King HA, Trotter KW, Archer TK. King HA, et al. Biochim Biophys Acta. 2012 Jul;1819(7):716-26. doi: 10.1016/j.bbagrm.2012.02.019. Epub 2012 Mar 6. Biochim Biophys Acta. 2012. PMID: 22425674 Free PMC article. Review.
Cited by
- Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation.
Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L. Letimier FA, et al. EMBO J. 2007 Mar 7;26(5):1292-302. doi: 10.1038/sj.emboj.7601586. Epub 2007 Feb 15. EMBO J. 2007. PMID: 17304212 Free PMC article. - Intrinsic Disorder of the BAF Complex: Roles in Chromatin Remodeling and Disease Development.
El Hadidy N, Uversky VN. El Hadidy N, et al. Int J Mol Sci. 2019 Oct 23;20(21):5260. doi: 10.3390/ijms20215260. Int J Mol Sci. 2019. PMID: 31652801 Free PMC article. - Differential Regulation of Type I and Type III Interferon Signaling.
Stanifer ML, Pervolaraki K, Boulant S. Stanifer ML, et al. Int J Mol Sci. 2019 Mar 21;20(6):1445. doi: 10.3390/ijms20061445. Int J Mol Sci. 2019. PMID: 30901970 Free PMC article. Review. - Proteomic survey of ubiquitin-linked nuclear proteins in interferon-stimulated macrophages.
Kim JY, Anderson ED, Huynh W, Dey A, Ozato K. Kim JY, et al. J Interferon Cytokine Res. 2011 Aug;31(8):619-28. doi: 10.1089/jir.2011.0006. Epub 2011 Mar 23. J Interferon Cytokine Res. 2011. PMID: 21428739 Free PMC article. - IRF9 is a key factor for eliciting the antiproliferative activity of IFN-alpha.
Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC. Tsuno T, et al. J Immunother. 2009 Oct;32(8):803-16. doi: 10.1097/CJI.0b013e3181ad4092. J Immunother. 2009. PMID: 19752753 Free PMC article.
References
- Aalfs, J. D., and R. E. Kingston. 2000. What does ′chromatin remodeling' mean? Trends Biochem. Sci. 25:548-555. - PubMed
- Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. - PubMed
- Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765-775. - PubMed
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. - PubMed
- Bhattacharya, S., R. Eckner, S. Grossman, E. Oldread, Z. Arany, A. D'Andrea, and D. M. Livingston. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature 383:344-347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous