Analysis of segmental duplications and genome assembly in the mouse - PubMed (original) (raw)
Analysis of segmental duplications and genome assembly in the mouse
Jeffrey A Bailey et al. Genome Res. 2004 May.
Abstract
Limited comparative studies suggest that the human genome is particularly enriched for recent segmental duplications. The extent of segmental duplications in other mammalian genomes is unknown and confounded by methodological differences in genome assembly. Here, we present a detailed analysis of recent duplication content within the mouse genome using a whole-genome assembly comparison method and a novel assembly independent method, designed to take advantage of the reduced allelic variation of the C57BL/6J strain. We conservatively estimate that approximately 57% of all highly identical segmental duplications (>or=90%) were misassembled or collapsed within the working draft WGS assembly. The WGS approach often leaves duplications fragmented and unassigned to a chromosome when compared with the clone-ordered-based approach. Our preliminary analysis suggests that 1.7%-2.0% of the mouse genome is part of recent large segmental duplications (about half of what is observed for the human genome). We have constructed a mouse segmental duplication database to aid in the characterization of these regions and their integration into the final mouse genome assembly. This work suggests significant biological differences in the architecture of recent segmental duplications between human and mouse. In addition, our unique method provides the means for improving whole-genome shotgun sequence assembly of mouse and future mammalian genomes.
Figures
Figure 1
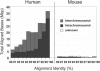
Whole-genome assembly comparison for mouse and human. We compared the sum of aligned bases (excluding gaps) for segmental duplications represented by alignments ≥10 kb in both the human genome (build 31) and the draft mouse genome (MGSCv3). Both the human and mouse genomes have alignments at all levels of identity; however, the human genome has a dramatically greater amount of aligned bases relative to the mouse (227,812 kbp vs. 10,042 kbp). The number of alignments increases geometrically relative to the number of copies. Mouse appears relatively rich in intrachromosomal duplications (black) and lacking in interchromosomal duplications (dark gray). However, many alignments are poorly characterized as indicated by the enrichment within the unplaced chromosome (chrUn—light gray).
Figure 2
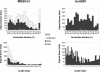
Whole-genome alignment (WGAC) statistics of the mouse draft and the build29 finished genome. Alignment statistics are binned in terms of percent identity or length (≥10 kb). We performed BLAST-based segmental duplication detection on MGSCv3 and the finished portion of build 29. The finished build 29 subset represents 439 Mb (17.7% of the draft assembly size). The abundance of aligned bases between 99.5%–100% that map to the unknown chromosome in MGSCv3 may represent highly similar duplication requiring further characterization. The build 29 pairwise were hand curated to remove uncharacterized interspersed transposable elements (Methods).
Figure 3
Examples of whole-genome shotgun sequence detection (WSSD). The calibration of our WSSD method was performed on a set of unique and duplicated sequences. Unique sequences were drawn from clones shown to be unique by both metaphase and interphase FISH (e.g., AL590991). Examples of duplicated sequence were drawn from recently described pericentromeric duplications (e.g., mmu5; Thomas et al. 2003). Detection parameters were optimized to differentiate unique from duplicated sequence. Black dots represent the similarity and position of individual sequence reads. Masked repetitive regions (LINE elements, purple; ERV elements, green; and simple sequence repeats, red) are shown as vertical bars. From previous studies of the human genome (Bailey et al. 2002a), read depth (blue line) provided the measure for duplication detection. Here, we also took advantage of the reduced level of allelic variation within the C57BL/6J strain to increase our power. Thus, single base-pair differences most likely signify either paralogous sequence or sequencing errors. By excluding errors (through the calculation of read identity using only high quality base positions), we could categorize each read as allelic (≥99.8% identity) or paralogous (<99.8% identity). Regions showing a divergent read ratio (red line) of >0.8 (paralogous: allelic) were deemed duplicated. A divergent read ratio of 1 would suggest one paralogous copy.
Figure 4
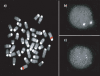
FISH confirmation. An example of (a) metaphase and (b) interphase FISH hybridization with a duplicated BAC clone (RP23–3D2; see Table 4) that was identified by the whole-genome shotgun detection strategy. Increased signal intensity was confirmed using (c) cohybridization with a unique probe (RP21-344N12) in the same nucleus as shown in b. Tandem segmental duplications were most frequently observed (Table 4). The results of all FISH experiments are available online (http://www.biologia.uniba.it/mouse/).
Figure 5
Mouse segmental duplications. Segmental duplications detected by whole-genome shotgun sequence detection (WSSD, black bars) and whole-genome analysis comparison (WGAC, red/blue bars) are drawn to scale within the published mouse genome assembly (MGSC 2002). Chromosome lengths and the centromere positions are shown in purple. These data are available as part of an interactive mouse segmental duplication database (
).
Similar articles
- Shotgun sequence assembly and recent segmental duplications within the human genome.
She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE. She X, et al. Nature. 2004 Oct 21;431(7011):927-30. doi: 10.1038/nature03062. Nature. 2004. PMID: 15496912 - Recent segmental and gene duplications in the mouse genome.
Cheung J, Wilson MD, Zhang J, Khaja R, MacDonald JR, Heng HH, Koop BF, Scherer SW. Cheung J, et al. Genome Biol. 2003;4(8):R47. doi: 10.1186/gb-2003-4-8-r47. Epub 2003 Jul 9. Genome Biol. 2003. PMID: 12914656 Free PMC article. - Recent segmental duplications in the working draft assembly of the brown Norway rat.
Tuzun E, Bailey JA, Eichler EE. Tuzun E, et al. Genome Res. 2004 Apr;14(4):493-506. doi: 10.1101/gr.1907504. Genome Res. 2004. PMID: 15059990 Free PMC article. - The value of new genome references.
Worley KC, Richards S, Rogers J. Worley KC, et al. Exp Cell Res. 2017 Sep 15;358(2):433-438. doi: 10.1016/j.yexcr.2016.12.014. Epub 2016 Dec 23. Exp Cell Res. 2017. PMID: 28017728 Free PMC article. Review. - Recent duplication, domain accretion and the dynamic mutation of the human genome.
Eichler EE. Eichler EE. Trends Genet. 2001 Nov;17(11):661-9. doi: 10.1016/s0168-9525(01)02492-1. Trends Genet. 2001. PMID: 11672867 Review.
Cited by
- Mouse segmental duplication and copy number variation.
She X, Cheng Z, Zöllner S, Church DM, Eichler EE. She X, et al. Nat Genet. 2008 Jul;40(7):909-14. doi: 10.1038/ng.172. Epub 2008 May 22. Nat Genet. 2008. PMID: 18500340 Free PMC article. - Dissecting a hidden gene duplication: the Arabidopsis thaliana SEC10 locus.
Vukašinović N, Cvrčková F, Eliáš M, Cole R, Fowler JE, Žárský V, Synek L. Vukašinović N, et al. PLoS One. 2014 Apr 11;9(4):e94077. doi: 10.1371/journal.pone.0094077. eCollection 2014. PLoS One. 2014. PMID: 24728280 Free PMC article. - Extended regions of suspected mis-assembly in the rat reference genome.
Ramdas S, Ozel AB, Treutelaar MK, Holl K, Mandel M, Woods LCS, Li JZ. Ramdas S, et al. Sci Data. 2019 Apr 23;6(1):39. doi: 10.1038/s41597-019-0041-6. Sci Data. 2019. PMID: 31015470 Free PMC article. - Evolutionary Dynamics of the POTE Gene Family in Human and Nonhuman Primates.
Maggiolini FAM, Mercuri L, Antonacci F, Anaclerio F, Calabrese FM, Lorusso N, L'Abbate A, Sorensen M, Giannuzzi G, Eichler EE, Catacchio CR, Ventura M. Maggiolini FAM, et al. Genes (Basel). 2020 Feb 18;11(2):213. doi: 10.3390/genes11020213. Genes (Basel). 2020. PMID: 32085667 Free PMC article. - The Taxus genome provides insights into paclitaxel biosynthesis.
Xiong X, Gou J, Liao Q, Li Y, Zhou Q, Bi G, Li C, Du R, Wang X, Sun T, Guo L, Liang H, Lu P, Wu Y, Zhang Z, Ro DK, Shang Y, Huang S, Yan J. Xiong X, et al. Nat Plants. 2021 Aug;7(8):1026-1036. doi: 10.1038/s41477-021-00963-5. Epub 2021 Jul 15. Nat Plants. 2021. PMID: 34267359 Free PMC article.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. - PubMed
- Armengol, L., Pujana, M.A., Cheung, J., Scherer, S.W., and Estivill, X. 2003. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 12: 2201-2208. - PubMed
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002a. Recent segmental duplications in the human genome. Science 297: 1003-1007. - PubMed
WEB SITE REFERENCES
- http://mouseparalogy.gene.cwru.edu; Eichler Lab Mouse Segmental Duplication Database.
- http://www.biologia.uniba.it/mouse/; FISH experiments of WSSD duplication-positive clones.
- http://www.ncbi.nlm.nih.gov/genome/guide/build.html; NCBI's Genome Annotation Pipeline.
- http://www.ncbi.nlm.nih.gov/genome/guide/mouse/MmStats.html; Mouse Build 30 Statistics.
- http://www.ncbi.nlm.nih.gov/RefSeq/; NCBI Reference Sequence Database.
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM058815/GM/NIGMS NIH HHS/United States
- GM58815/GM/NIGMS NIH HHS/United States
- HG002385/HG/NHGRI NIH HHS/United States
- C.50/TI_/Telethon/Italy
- CA094816/CA/NCI NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 HG002385/HG/NHGRI NIH HHS/United States
- T32 GM007250/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources