RNA surveillance down-regulates expression of nonfunctional kappa alleles and detects premature termination within the last kappa exon - PubMed (original) (raw)
RNA surveillance down-regulates expression of nonfunctional kappa alleles and detects premature termination within the last kappa exon
Laurent Delpy et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Random V(D)J junctions would generate nonfunctional and/or out-of-frame sequences in about two-thirds of cases and result in abundant transcripts encoding truncated proteins. Although allelic exclusion at the DNA recombination level ensures that a single allele is functional, the frequent biallelic rearrangements need additional mechanisms to down-regulate aberrant transcripts in those cells with both a functionally and a nonfunctionally rearranged allele. The process of nonsense-mediated decay targets aberrantly rearranged Ig heavy-chain transcripts, but the situation of light-chain mRNAs is more complex, because they do not meet the usual requirements for nonsense-mediated decay and most often lack a spliceable intron downstream of the premature termination. We studied immunoglobulin heavy-chain -/- pro-B cells in which light chain genes get rearranged and expressed in the absence of any selection for the assembly of a functional B cell receptor. Using this model, we show that the whole kappa locus is accessible in pro-B cells and allows the assembly of a broad spectrum of VkappaJkappa segments, most of which are out-of-frame. This model provides an evaluation of the in vivo efficiency of RNA surveillance toward aberrant kappa mRNAs produced in pro-B cells. Our data show that nonfunctional kappa transcripts are excluded from the mature mRNA pool not only by detecting termination in an upstream exon but also by detecting changes in the position of termination within the last exon. Similar mechanisms efficiently down-regulate nonfunctional kappa transcripts arising in normal mature B cells due to the biallelic transcription of rearranged kappa genes.
Figures
Fig. 1.
Vκ-Jκ junctions in bone marrow pro-B cells from N/N homozygous mice. Sequences are given with reference to the name of the corresponding germ-line Vκ and Jκ segment. Dashes indicate junctional deletions; N insertions (bold) and P insertions are indicated in between. Italicized nucleotides encode the CDR3 segment (whose length is indicated in parentheses); nonfunctional sequences are marked by an asterisk with indication of the +1 or +2 frameshift or of an in-frame stop codon (in upper case).
Fig. 2.
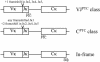
Schematic structure of the various classes of Vκ-Jκ rearrangements. Positions of the PTC resulting from a frameshifted rearrangement and of the normal in-frame stop codon are indicated (***).
Fig. 3.
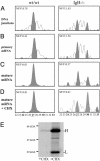
CDR3 lengths and out-of-frame κ junctions. Bone marrow samples from either WT/WT or IgH-/- animals blocked at the pro-B cell stage were analyzed by high-resolution capillary electrophoresis. Data from a representative experiment are shown at the DNA level (A), the primary RNA level (B), and the mature RNA level [without cycloheximide (C) and with cycloheximide (D)]. Peaks corresponding to in-frame rearrangements are colored gray (including the dominant CDR3 with a canonical length of 27 nt); NF/F ratios are indicated. (E) Control of protein synthesis inhibition in cycloheximide-treated vs. untreated splenocyte cultures (metabolically labeled Ig were adsorbed on protein G and separated on a polyacrylamide gel).
Fig. 4.
Relative frequencies of out-of-frame κ junctions. NF/F ratios were estimated at the DNA, primary transcript, and mature mRNA levels by multiple experiments of capillary electrophoresis in IgH-/- pro-B cells or in tissues from normal mice (bone marrow, spleen, and κ-expressing sorted splenocytes). Filled diamonds represent values determined for Vκ-Jκ1 junctions in DNA and primary RNA and for Vκ-Jκ-Cκ junctions in mature transcripts; open squares represent Vκ-Jκ5 junctions in DNA and primary RNA.
Similar articles
- Multiple RNA surveillance mechanisms cooperate to reduce the amount of nonfunctional Ig kappa transcripts.
Chemin G, Tinguely A, Sirac C, Lechouane F, Duchez S, Cogné M, Delpy L. Chemin G, et al. J Immunol. 2010 May 1;184(9):5009-17. doi: 10.4049/jimmunol.0902949. Epub 2010 Mar 31. J Immunol. 2010. PMID: 20357261 - Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion.
Kitamura D, Rajewsky K. Kitamura D, et al. Nature. 1992 Mar 12;356(6365):154-6. doi: 10.1038/356154a0. Nature. 1992. PMID: 1545868 - The Yin and Yang of RNA surveillance in B lymphocytes and antibody-secreting plasma cells.
Lambert JM, Srour N, Delpy L. Lambert JM, et al. BMB Rep. 2019 Dec;52(12):671-678. doi: 10.5483/BMBRep.2019.52.12.232. BMB Rep. 2019. PMID: 31619318 Free PMC article. Review.
Cited by
- A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay.
Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. Behm-Ansmant I, et al. EMBO J. 2007 Mar 21;26(6):1591-601. doi: 10.1038/sj.emboj.7601588. Epub 2007 Feb 22. EMBO J. 2007. PMID: 17318186 Free PMC article. - Synonymous codon usage affects the expression of wild type and F508del CFTR.
Shah K, Cheng Y, Hahn B, Bridges R, Bradbury NA, Mueller DM. Shah K, et al. J Mol Biol. 2015 Mar 27;427(6 Pt B):1464-1479. doi: 10.1016/j.jmb.2015.02.003. Epub 2015 Feb 10. J Mol Biol. 2015. PMID: 25676312 Free PMC article. - Mechanisms and Regulation of Nonsense-Mediated mRNA Decay and Nonsense-Associated Altered Splicing in Lymphocytes.
Lambert JM, Ashi MO, Srour N, Delpy L, Saulière J. Lambert JM, et al. Int J Mol Sci. 2020 Feb 17;21(4):1335. doi: 10.3390/ijms21041335. Int J Mol Sci. 2020. PMID: 32079193 Free PMC article. Review. - Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development.
Tinguely A, Chemin G, Péron S, Sirac C, Reynaud S, Cogné M, Delpy L. Tinguely A, et al. Mol Cell Biol. 2012 Jan;32(1):107-17. doi: 10.1128/MCB.06138-11. Epub 2011 Oct 28. Mol Cell Biol. 2012. PMID: 22037763 Free PMC article. - Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing.
Baek D, Green P. Baek D, et al. Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12813-8. doi: 10.1073/pnas.0506139102. Epub 2005 Aug 25. Proc Natl Acad Sci U S A. 2005. PMID: 16123126 Free PMC article.
References
- Coleclough, C., Perry, R. P., Karjalainen, K. & Weigert, M. (1981) Nature 290, 372-378. - PubMed
- Victor, K. D., Vu, K. & Feeney, A. J. (1994) J. Immunol. 152, 3467-3475. - PubMed
- Arakawa, H., Shimizu, T. & Takeda, S. (1996) Int. Immunol. 8, 91-99. - PubMed
- Melchers, F., ten Boekel, E., Yamagami, T., Andersson, J. & Rolink, A. (1999) Semin. Immunol. 11, 307-317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources