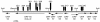Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2 - PubMed (original) (raw)
Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2
Larry D Hodges et al. J Bacteriol. 2004 May.
Abstract
Agrobacterium tumefaciens and Agrobacterium rhizogenes transfer plasmid-encoded genes and virulence (Vir) proteins into plant cells. The transferred DNA (T-DNA) is stably inherited and expressed in plant cells, causing crown gall or hairy root disease. DNA transfer from A. tumefaciens into plant cells resembles plasmid conjugation; single-stranded DNA (ssDNA) is exported from the bacteria via a type IV secretion system comprised of VirB1 through VirB11 and VirD4. Bacteria also secrete certain Vir proteins into plant cells via this pore. One of these, VirE2, is an ssDNA-binding protein crucial for efficient T-DNA transfer and integration. VirE2 binds incoming ssT-DNA and helps target it into the nucleus. Some strains of A. rhizogenes lack VirE2, but they still transfer T-DNA efficiently. We isolated a novel gene from A. rhizogenes that restored pathogenicity to virE2 mutant A. tumefaciens. The GALLS gene was essential for pathogenicity of A. rhizogenes. Unlike VirE2, GALLS contains a nucleoside triphosphate binding motif similar to one in TraA, a strand transferase conjugation protein. Despite their lack of similarity, GALLS substituted for VirE2.
Figures
FIG. 1.
(A to H) Carrots inoculated on the basal surface with derivatives of the virE2 mutant A. tumefaciens MX358 containing pRi1724::kan (A), pLH77 (the GALLS gene-positive cosmid) (B), pLH77-mutant GALLS gene containing Tn_3-lac-301_ (C), pLH77-mutant GALLS gene containing Tn_3-lac-321_ (D), pLH338 (the GALLS gene-positive plasmid) (E), pLH345 (mutant GALLS gene with deletion of _Eco_RI plasmid) (F), pLH344 (mutant GALLS gene with deletion of _Nco_I plasmid) (G), pVK100 (cosmid-plasmid vector) (H). (I to K) Mixed infections (of the basal surface) with C58C1(pRi1724::kan) plus MX358 (virE2 mutant) (I), MX234(pRi1724::kan) (virB1 mutant) plus MX358 (J), and MX368(pRi1724::kan) (virB10 mutant) plus MX358 (K). (L) Basal surface of a carrot inoculated with wild-type A. tumefaciens A348. (M to P) Uninoculated basal (M) and apical (N) surfaces and carrots inoculated on the apical surface with A. rhizogenes MX599 (O) and A. rhizogenes K599 (P). MX599 is a Tn_3-lac-301_-containing mutant GALLS gene derivative of wild-type A. rhizogenes K599 (Table 1).
FIG. 2.
Tn_3-lac_ insertions in the GALLS gene region. The map shows the 17,437-bp region shared by seven cosmids that complemented mutations in virE2. The arrows beneath the map indicate the locations and orientations of ORFs. The vertical arrows represent Tn_3-lac_ insertions; allele numbers are above each arrow. The short vertical arrows indicate Tn_3-lac_ insertions oriented with the lac operon in the sense orientation relative to the disrupted coding sequence; all of the insertions are transcriptional (out-of-frame) fusions of the ORF with the lac genes, except for allele 335, which formed a translational (in-frame) fusion with the GALLS gene. The long vertical arrows indicate Tn_3-lac_ insertions in which the lac genes are oriented antisense relative to the coding sequence.
FIG. 3.
Domains in the GALLS protein. Conserved NTP-binding/helicase motifs (I, II, and III), TraA-like sequences (1 to 5), a putative NLS, and GALLS repeats are shown. _Nco_I, restriction sites used to make in-frame deletions within the GALLS repeats (short underlined region); _Eco_RI, restriction site used to remove the entire 3′ end of the GALLS gene, including the NLS, GALLS repeats, and C terminus (long underlined region).
FIG. 4.
The amino acid sequences of the helicase domains of TraA encoded by pTiC58 and pRi1724 were aligned with the corresponding regions of GALLS encoded by pRi1724 and pRiA4. We used the ClustalW program to align the sequences. ATP-binding domains identified by Farrand et al. (21) are underlined with a dashed line. The numbers indicate the locations of the adjacent amino acid in each protein. Solid boxes, amino acids that are identical in all four proteins; shaded boxes, similar amino acids. The groups of amino acids considered similar in this analysis were I, L, M, and V; A, G, and S; H, K, and R; D and E; N and Q; F, W, and Y; and S and T. Dashes indicate gaps placed in the sequences by the ClustalW program to maximize alignment.
FIG. 5.
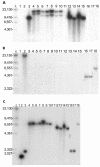
Southern blot analysis of the GALLS, virE2, and virD2 genes in various A. rhizogenes, A. tumefaciens, and A. vitis strains. The blots were probed with radiolabeled GALLS gene (A), virE2 (B), or virD2 (C) sequences. λ, phage λ DNA digested with HindIII. The size of each band in base pairs is indicated. Each panel contained DNA from the following strains: lanes 1, A136 (no Ti plasmid); lanes 2, A348 (octopine-type pTiA6 in A136); lanes 3, C58C1(pRi1724::kan) (mikimopine-type Ri plasmid); lanes 4, A136(pRiA4) (agropine-type Ri plasmid); lanes 5, C58C1(pRiA4) (agropine-type Ri plasmid); lanes 6, ATCC 15834 (agropine-type A. rhizogenes); lanes 7, A4 (no. 117) (agropine-type A. rhizogenes A4; isolate 117); lanes 8, A4 (no. 100) (agropine-type A. rhizogenes A4; isolate 100); lanes 9, NCPPB1855 (agropine-type A. rhizogenes); lanes 10, C58C1RS(pRiTR105) (agropine-type Ri plasmid); lanes 11, C58C1RS(pArA4a) (large plasmid [not associated with virulence] from A. rhizogenes A4); lanes 12, NCPPB2655 (cucumopine-type A. rhizogenes); lanes 13, NCPPB2657 (cucumopine-type A. rhizogenes); lanes 14, NCPPB2659 (cucumopine-type A. rhizogenes K599); lanes 15, NT1(pRiK599) (cucumopine-type Ri plasmid); lanes 16, NCIB8196 (mannopine-type A. rhizogenes); lanes 17, ICPB-TR7 (mannopine-type A. rhizogenes); lanes 18, A. vitis A856 (limited-host-range grape-specific strain).
FIG. 6.
Arrangement of virulence (vir) operons in octopine-type (pTiA6) (A), nopaline-type (pTiC58) (B), and mikimopine-type (pRi1724) (C) plasmids.
Similar articles
- The Agrobacterium rhizogenes GALLS gene encodes two secreted proteins required for genetic transformation of plants.
Hodges LD, Lee LY, McNett H, Gelvin SB, Ream W. Hodges LD, et al. J Bacteriol. 2009 Jan;191(1):355-64. doi: 10.1128/JB.01018-08. Epub 2008 Oct 24. J Bacteriol. 2009. PMID: 18952790 Free PMC article. - Agrobacterium rhizogenes GALLS protein contains domains for ATP binding, nuclear localization, and type IV secretion.
Hodges LD, Vergunst AC, Neal-McKinney J, den Dulk-Ras A, Moyer DM, Hooykaas PJ, Ream W. Hodges LD, et al. J Bacteriol. 2006 Dec;188(23):8222-30. doi: 10.1128/JB.00747-06. Epub 2006 Sep 29. J Bacteriol. 2006. PMID: 17012398 Free PMC article. - Agrobacterium tumefaciens and A. rhizogenes use different proteins to transport bacterial DNA into the plant cell nucleus.
Ream W. Ream W. Microb Biotechnol. 2009 Jul;2(4):416-27. doi: 10.1111/j.1751-7915.2009.00104.x. Epub 2009 Apr 6. Microb Biotechnol. 2009. PMID: 21255274 Free PMC article. Review. - pSa causes oncogenic suppression of Agrobacterium by inhibiting VirE2 protein export.
Lee LY, Gelvin SB, Kado CI. Lee LY, et al. J Bacteriol. 1999 Jan;181(1):186-96. doi: 10.1128/JB.181.1.186-196.1999. J Bacteriol. 1999. PMID: 9864329 Free PMC article. - The VirE2 protein of Agrobacterium tumefaciens: the Yin and Yang of T-DNA transfer.
Duckely M, Hohn B. Duckely M, et al. FEMS Microbiol Lett. 2003 Jun 6;223(1):1-6. doi: 10.1016/S0378-1097(03)00246-5. FEMS Microbiol Lett. 2003. PMID: 12798992 Review.
Cited by
- The genome sequence of hairy root Rhizobium rhizogenes strain LBA9402: Bioinformatics analysis suggests the presence of a new opine system in the agropine Ri plasmid.
Hooykaas MJG, Hooykaas PJJ. Hooykaas MJG, et al. Microbiologyopen. 2021 Mar;10(2):e1180. doi: 10.1002/mbo3.1180. Microbiologyopen. 2021. PMID: 33970547 Free PMC article. - Traversing the Cell: Agrobacterium T-DNA's Journey to the Host Genome.
Gelvin SB. Gelvin SB. Front Plant Sci. 2012 Mar 26;3:52. doi: 10.3389/fpls.2012.00052. eCollection 2012. Front Plant Sci. 2012. PMID: 22645590 Free PMC article. - The Agrobacterium rhizogenes GALLS gene encodes two secreted proteins required for genetic transformation of plants.
Hodges LD, Lee LY, McNett H, Gelvin SB, Ream W. Hodges LD, et al. J Bacteriol. 2009 Jan;191(1):355-64. doi: 10.1128/JB.01018-08. Epub 2008 Oct 24. J Bacteriol. 2009. PMID: 18952790 Free PMC article. - New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation.
Pitzschke A, Hirt H. Pitzschke A, et al. EMBO J. 2010 Mar 17;29(6):1021-32. doi: 10.1038/emboj.2010.8. Epub 2010 Feb 11. EMBO J. 2010. PMID: 20150897 Free PMC article. Review. - The heterologous expression of conserved Glycine max (soybean) mitogen activated protein kinase 3 (MAPK3) paralogs suppresses Meloidogyne incognita parasitism in Gossypium hirsutum (upland cotton).
Klink VP, Alkharouf NW, Lawrence KS, Lawaju BR, Sharma K, Niraula PM, McNeece BT. Klink VP, et al. Transgenic Res. 2022 Oct;31(4-5):457-487. doi: 10.1007/s11248-022-00312-y. Epub 2022 Jun 28. Transgenic Res. 2022. PMID: 35763120 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources