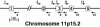Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase - PubMed (original) (raw)
Genetic evidence that the human CYP2R1 enzyme is a key vitamin D 25-hydroxylase
Jeffrey B Cheng et al. Proc Natl Acad Sci U S A. 2004.
Abstract
The synthesis of bioactive vitamin D requires hydroxylation at the 1 alpha and 25 positions by cytochrome P450 enzymes in the kidney and liver, respectively. The mitochondrial enzyme CYP27B1 catalyzes 1 alpha-hydroxylation in the kidney but the identity of the hepatic 25-hydroxylase has remained unclear for >30 years. We previously identified the microsomal CYP2R1 protein as a potential candidate for the liver vitamin D 25-hydroxylase based on the enzyme's biochemical properties, conservation, and expression pattern. Here, we report a molecular analysis of a patient with low circulating levels of 25-hydroxyvitamin D and classic symptoms of vitamin D deficiency. This individual was found to be homozygous for a transition mutation in exon 2 of the CYP2R1 gene on chromosome 11p15.2. The inherited mutation caused the substitution of a proline for an evolutionarily conserved leucine at amino acid 99 in the CYP2R1 protein and eliminated vitamin D 25-hydroxylase enzyme activity. These data identify CYP2R1 as a biologically relevant vitamin D 25-hydroxylase and reveal the molecular basis of a human genetic disease, selective 25-hydroxyvitamin D deficiency.
Figures
Fig. 1.
Structure of the human CYP2R1 gene. The predicted exon–intron structure of the human CYP2R1 gene is shown together with six pairs of oligonucleotides used to amplify and sequence fragments of the DNA. Exon 3 was amplified with primer pair 5, 8, and was sequenced with primers 5, 6, 7, and 8. The gene is drawn to scale with the exception of intron 1, which is estimated to be 9.1 kb in length. The autosomal location of the gene was deduced from the Human Genome database, which can be accessed at
www.ncbi.nlm.nih.gov/mapview/maps.cgi?taxid=9606&chr=11
.
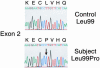
Fig. 2.
DNA sequence analysis of mutation in CYP2R1 vitamin D 25-hydroxylase. The partial DNA sequence of exon 2 from the CYP2R1 gene is shown from a normal individual (Upper) and from an individual with selective 25-hydroxyvitamin D deficiency (Lower). The codon specifying amino acid 99 is C_T_T in the normal gene and C_C_T (arrow) in the affected individual. The T → C transition mutation in the second nucleotide causes a change from leucine to proline at residue 99. All other nucleotides in the CYP2R1 gene of the proband were identical to those in normal individuals.
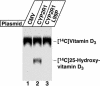
Fig. 3.
Biochemical assay of CYP2R1 vitamin D 25-hydroxylase enzyme activity. HEK 293 cells were transfected with the indicated expression plasmids for a period of 18–20 h. Thereafter, the medium was made 4.6 × 10-7 M in [4-14C]vitamin D3 and the incubation continued for an additional 96 h. Lipids were extracted from cells and medium into chloroform:methanol (2:1, vol/vol), and vitamin D metabolites and standards were separated by TLC on 150-Å silica gel plates (Whatman, catalog no. 4855–821) in a solvent system containing cyclohexane:ethyl acetate (3:2, vol/vol). After development, radioactivity was detected by PhosphorImager analysis, and the positions to which authentic vitamin D3 and 25-hydroxyvitamin D3 migrated on the plate were determined by staining with iodine.
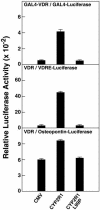
Fig. 4.
L99P mutation in CYP2R1 causes loss of vitamin D3 activation. Expression vectors encoding no protein, the normal CYP2R1 enzyme (CYP2R1), or a version containing a proline substituted for leucine at amino acid 99 (CYP2R1L99P), were transfected in triplicate into HEK 293 cells together with the indicated VDR-reporter gene systems. The concentrations of vitamin D3 added to the medium ranged from 0.25 to 1.0 μM. After 16–20 h, cells were lysed and assayed for enzyme activities. Relative LUC activity was calculated by dividing units of LUC enzyme activity measured on a Dynex MLX luminometer by units of β-galactosidase enzyme activity measured on a Dynex Opsys MR spectrophotometer. Means ± SE of measurement were calculated and plotted in histogram form. The data are representative of two separate experiments carried out on different days.
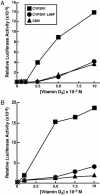
Fig. 5.
Response of normal and L99P CYP2R1 enzymes to increasing concentrations of vitamin D3 (A) and vitamin D2 (B). The indicated expression plasmids were introduced into HEK 293 cells together with DNAs constituting the VDR/VDRE-LUC reporter gene system. After an expression period (8 h), different amounts of the two forms of vitamin D were added to the medium and the incubation was continued for an additional 16 h. Thereafter, cells were lysed and assayed for LUC and β-galactosidase enzyme activities. Points on the graphs represent means of triplicate values established at each concentration of secosteroid. These experiments were repeated at least two times each.
Similar articles
- De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxilase.
Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. Cheng JB, et al. J Biol Chem. 2003 Sep 26;278(39):38084-93. doi: 10.1074/jbc.M307028200. Epub 2003 Jul 16. J Biol Chem. 2003. PMID: 12867411 Free PMC article. - Mutation of the CYP2R1 vitamin D 25-hydroxylase in a Saudi Arabian family with severe vitamin D deficiency.
Al Mutair AN, Nasrat GH, Russell DW. Al Mutair AN, et al. J Clin Endocrinol Metab. 2012 Oct;97(10):E2022-5. doi: 10.1210/jc.2012-1340. Epub 2012 Aug 1. J Clin Endocrinol Metab. 2012. PMID: 22855339 Free PMC article. - Vitamin D hydroxylases CYP2R1, CYP27B1 and CYP24A1 in renal cell carcinoma.
Urbschat A, Paulus P, von Quernheim QF, Brück P, Badenhoop K, Zeuzem S, Ramos-Lopez E. Urbschat A, et al. Eur J Clin Invest. 2013 Dec;43(12):1282-90. doi: 10.1111/eci.12176. Epub 2013 Oct 12. Eur J Clin Invest. 2013. PMID: 24245571 - CYP2R1 mutations causing vitamin D-deficiency rickets.
Thacher TD, Levine MA. Thacher TD, et al. J Steroid Biochem Mol Biol. 2017 Oct;173:333-336. doi: 10.1016/j.jsbmb.2016.07.014. Epub 2016 Jul 27. J Steroid Biochem Mol Biol. 2017. PMID: 27473561 Review. - Cytochrome P450 enzymes in the bioactivation of vitamin D to its hormonal form (review).
Wikvall K. Wikvall K. Int J Mol Med. 2001 Feb;7(2):201-9. doi: 10.3892/ijmm.7.2.201. Int J Mol Med. 2001. PMID: 11172626 Review.
Cited by
- Vitamin D: not just the bone. Evidence for beneficial pleiotropic extraskeletal effects.
Caprio M, Infante M, Calanchini M, Mammi C, Fabbri A. Caprio M, et al. Eat Weight Disord. 2017 Mar;22(1):27-41. doi: 10.1007/s40519-016-0312-6. Epub 2016 Aug 23. Eat Weight Disord. 2017. PMID: 27553017 Review. - Hereditary Rickets: A Quick Guide for the Pediatrician.
AlSubaihin A, Harrington J. AlSubaihin A, et al. Curr Pediatr Rev. 2024;20(4):380-394. doi: 10.2174/1573396319666221205123402. Curr Pediatr Rev. 2024. PMID: 36475338 Review. - Lucky, times ten: A career in Texas science.
Russell DW. Russell DW. J Biol Chem. 2018 Dec 7;293(49):18804-18827. doi: 10.1074/jbc.X118.005918. J Biol Chem. 2018. PMID: 30530852 Free PMC article. - Vitamin D deficiency and toxicity in chronic kidney disease: in search of the therapeutic window.
Querfeld U, Mak RH. Querfeld U, et al. Pediatr Nephrol. 2010 Dec;25(12):2413-30. doi: 10.1007/s00467-010-1574-2. Epub 2010 Jun 22. Pediatr Nephrol. 2010. PMID: 20567854 Review. - 100 YEARS OF VITAMIN D: Historical aspects of vitamin D.
Jones G. Jones G. Endocr Connect. 2022 Apr 22;11(4):e210594. doi: 10.1530/EC-21-0594. Endocr Connect. 2022. PMID: 35245207 Free PMC article. Review.
References
- Jones, G., Strugnell, S. A. & DeLuca, H. F. (1998) Physiol. Rev. 78, 1193-1231. - PubMed
- Gupta, R. P., Hollis, B. W., Patel, S. B., Patrick, K. S. & Bell, N. H. (2004) J. Bone Miner. Res. 19, 680-688. - PubMed
- Yamasaki, T., Izumi, S., Ide, H. & Ohyama, Y. (March 16, 2004) J. Biol. Chem., 10.1074/jbc.M312601200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM 08014/GM/NIGMS NIH HHS/United States
- P01 HL020948/HL/NHLBI NIH HHS/United States
- T32 GM008014/GM/NIGMS NIH HHS/United States
- R01 DK056603/DK/NIDDK NIH HHS/United States
- HL 20948/HL/NHLBI NIH HHS/United States
- DK 56603/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases