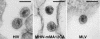Capsid is a dominant determinant of retrovirus infectivity in nondividing cells - PubMed (original) (raw)
Capsid is a dominant determinant of retrovirus infectivity in nondividing cells
Masahiro Yamashita et al. J Virol. 2004 Jun.
Abstract
A major difference between lentiviruses such as human immunodeficiency virus (HIV) and most other retroviruses is their ability to productively infect nondividing cells. We present here genetic evidence for involvement of the capsid protein (CA) in the infectious phenotype in nondividing cells. A chimeric HIV type 1 (HIV-1) in which the MA and CA of HIV-1 are replaced with the MA, p12, and CA encoding sequences from murine leukemia virus (MLV) loses the ability to efficiently infect nondividing cells. Analysis of the accumulation of two-long-terminal-repeat circles implies that the impairment of nuclear transport of preintegration complexes is responsible for the restricted infection of this chimeric virus in nondividing cells. Incorporation of MLV MA and MLV p12 into HIV virions alone does not exert any adverse effects on viral infection in interphase cells. These results suggest that CA is the dominant determinant for the difference between HIV and MLV in the ability to transduce nondividing cells.
Figures
FIG. 1.
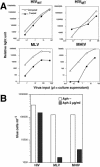
Generation of the infectious chimeric virus MHIV-mMA12CA. (A) Schematic representation of genomic organization of HIV-1, MLV, and MHIV (upper). Protein structure of the gag gene of these viruses is also shown (lower). MHIV-mMAp12CA encodes the MLV MA, p12, and CA and all the HIV-1 genes other than the HIV MA and CA. (B) Western blot analysis of purified virus particles of MHIV-mMA12CA together with HIV-1 and MLV. Polyprotein processing was tested for HIV-1 MA (p17gag) and CA (p24gag) as well as MLV MA (p15gag) and CA (p30gag) in addition to HIV-1 IN. Some unprocessed Gag and Gag-Pol proteins remain in the lane of MHIV-mMA12CA, but the anti-IN profile of MHIV-mMA12CA is the same as that of parental HIV-1 (not shown in this figure). (C) Single-cycle infectivity of MHIV-mMA12CA. Single-cycle infections of MHIV-mMA12CA were tested either alone or with envelope expression of VSV-G or a truncated HIV-1 Env in trans. Infectivity was measured with the MAGI assay by counting β-galactosidase-positive cells 2 days postinfection using viruses harvested after transfection of proviral clones with or without env expression vectors into 293T cells. These data are representative of three independent experiments.
FIG. 2.
Electron microscopy analysis of virions of HIV (left), MHIV-mMA12CA (center), and MLV (right) produced from 293T cells transfected with the respective proviruses. Bar, 100 nm.
FIG. 3.
MHIV-mMA12CA does not establish efficient infection in nondividing cells. (A) Infectivity of MHIV in aphidicolin-treated cells. MAGI cells were treated in the presence or absence of aphidicolin and challenged with virus stocks. Virus titers are expressed as the number of β-galactosidase-expressing blue cells per milliliter. These data are representative of five independent experiments. (B) Infectivity of MHIV in γ-irradiated cells. GHOST cells were treated with 3,500 rads from a 137cesium source. HIV and concentrated MHIV, all of which were pseudotyped with the VSV-G envelope protein, were spinoculated into normal (dividing) or γ-irradiated (nondividing) GHOST cells. Two days after infection, the cells were fixed and analyzed with flow cytometry for GFP-expressing GHOST cells. Dotted lines indicate zidovudine-treated control cells. (C) Macrophage infection with MHIV-mMA12CA. Monocyte-derived macrophages were prepared by adherence to the bottom of wells and maintained for 10 to 14 days before infection. Infectivity is shown as the relative percentage of luciferase titer in HeLa cells per that in macrophages. Error bars indicate standard deviations of duplicate cultures. The luciferase titers of these particular data are as follows: 534,677 relative light units (RLU) for HeLa cells infected with HIV; 680,954 RLU for HeLa cells infected with MLV; 137,651 RLU for HeLa cells infected with MHIV; 14,669 RLU for macrophages infected with HIV; 2,281 RLU for macrophages infected with MLV; 178 RLU for macrophages infected with MHIV. These values are averages of duplicate wells. The data presented here are representative of at least four different independent experiments using different blood donors.
FIG. 4.
(A) Dose-independent restriction of MHIV-mMA12CA in nondividing cells. Aphidicolin-treated HeLa cells were infected with increasing amount of luciferase-encoding viruses. Culture supernatants of transfected cells were used as inocula. Virus infectivity was judged by measuring luciferase titers of infected cell lysates 2 days after infection. (B) Transduction of the lacZ gene into dividing and nondividing HeLa cells. MAGI cells were treated in the presence or absence of aphidicolin and challenged with virus stocks by using retroviral vectors encoding the lacZ gene. Virus titers are expressed as the number of β-galactosidase-expressing blue cells per milliliter. These data are representative of two independent experiments.
FIG. 5.
CA determines infectivity in nondividing cells. (A) Schematic illustration of the protein structure of Gag of new chimeric viruses. Note that two new chimeric viruses as well as MHIV-mMA12CA were created based on the HIV-1 infectious clone. (B) Western blot analysis of purified virus particles of new MHIV chimeras. See the legend to Fig. 1B for details. (C) Single-cycle infectivity of MHIV as well as parental MLV and HIV. For details, see the legend to Fig. 1C.
FIG. 6.
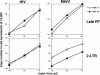
Infection of MHIV-mMA12CA is blocked at nuclear transport. Reverse transcription and nuclear transport of MHIV-mMA12CA and parental HIV-1 were monitored by measuring late reverse transcription products (upper) and 2-LTR circles (lower) as markers, respectively. Increasing amounts of virus inocula (x axis) were challenged with either normal or nondividing cells. The y axis indicates the copy number of each DNA analyzed with real-time quantitative PCR.
Similar articles
- Cooperative effect of gag proteins p12 and capsid during early events of murine leukemia virus replication.
Lee SK, Nagashima K, Hu WS. Lee SK, et al. J Virol. 2005 Apr;79(7):4159-69. doi: 10.1128/JVI.79.7.4159-4169.2005. J Virol. 2005. PMID: 15767417 Free PMC article. - Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells.
Qi M, Yang R, Aiken C. Qi M, et al. J Virol. 2008 Dec;82(24):12001-8. doi: 10.1128/JVI.01518-08. Epub 2008 Oct 1. J Virol. 2008. PMID: 18829762 Free PMC article. - Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells.
Chen BK, Rousso I, Shim S, Kim PS. Chen BK, et al. Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):15239-44. doi: 10.1073/pnas.261563198. Epub 2001 Dec 11. Proc Natl Acad Sci U S A. 2001. PMID: 11742097 Free PMC article. - Functional exchange of an oncoretrovirus and a lentivirus matrix protein.
Deminie CA, Emerman M. Deminie CA, et al. J Virol. 1994 Jul;68(7):4442-9. doi: 10.1128/JVI.68.7.4442-4449.1994. J Virol. 1994. PMID: 8207817 Free PMC article. - Determinants for lentiviral infection of non-dividing cells.
Vodicka MA. Vodicka MA. Somat Cell Mol Genet. 2001 Nov;26(1-6):35-49. doi: 10.1023/a:1021022629126. Somat Cell Mol Genet. 2001. PMID: 12465461 Review.
Cited by
- Identification of a genomic reservoir for new TRIM genes in primate genomes.
Han K, Lou DI, Sawyer SL. Han K, et al. PLoS Genet. 2011 Dec;7(12):e1002388. doi: 10.1371/journal.pgen.1002388. Epub 2011 Dec 1. PLoS Genet. 2011. PMID: 22144910 Free PMC article. - The Role of TNPO3 in HIV-1 Replication.
Diaz-Griffero F. Diaz-Griffero F. Mol Biol Int. 2012;2012:868597. doi: 10.1155/2012/868597. Epub 2012 Jul 19. Mol Biol Int. 2012. PMID: 22888429 Free PMC article. - Role of Transportin-SR2 in HIV-1 Nuclear Import.
Tabasi M, Nombela I, Janssens J, Lahousse AP, Christ F, Debyser Z. Tabasi M, et al. Viruses. 2021 May 4;13(5):829. doi: 10.3390/v13050829. Viruses. 2021. PMID: 34064404 Free PMC article. Review. - tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes.
Zaitseva L, Myers R, Fassati A. Zaitseva L, et al. PLoS Biol. 2006 Oct;4(10):e332. doi: 10.1371/journal.pbio.0040332. PLoS Biol. 2006. PMID: 17020411 Free PMC article. - Optimal cytoplasmic transport in viral infections.
D'Orsogna MR, Chou T. D'Orsogna MR, et al. PLoS One. 2009 Dec 30;4(12):e8165. doi: 10.1371/journal.pone.0008165. PLoS One. 2009. PMID: 20046829 Free PMC article.
References
- Alin, K., and S. P. Goff. 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology 222:339-351. - PubMed
- Balliet, J. W., D. L. Kolson, G. Eiger, F. M. Kim, K. A. McGann, A. Srinivasan, and R. Collman. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623-631. - PubMed
- Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. - PubMed
- Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI051153/AI/NIAID NIH HHS/United States
- R37 AI030927/AI/NIAID NIH HHS/United States
- R01 AI51153/AI/NIAID NIH HHS/United States
- R37 AI30927/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources