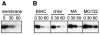Cell-cell fusion induced by the avian reovirus membrane fusion protein is regulated by protein degradation - PubMed (original) (raw)
Cell-cell fusion induced by the avian reovirus membrane fusion protein is regulated by protein degradation
Maya Shmulevitz et al. J Virol. 2004 Jun.
Abstract
The p10 fusion-associated small transmembrane protein of avian reovirus induces extensive syncytium formation in transfected cells. Here we show that p10-induced cell-cell fusion is restricted by rapid degradation of the majority of newly synthesized p10. The small ectodomain of p10 targets the protein for degradation following p10 insertion into an early membrane compartment. Paradoxically, conservative amino acid substitutions in the p10 ectodomain hydrophobic patch that eliminate fusion activity also increase p10 stability. The small amount of p10 that escapes intracellular degradation accumulates at the cell surface in a relatively stable form, where it mediates cell-cell fusion as a late-stage event in the virus replication cycle. The unusual relationship between a nonstructural viral membrane fusion protein and the replication cycle of a nonenveloped virus has apparently contributed to the evolution of a novel mechanism for restricting the extent of virus-induced cell-cell fusion.
Figures
FIG. 1.
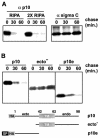
The ectodomain targets p10 for rapid degradation. (A) (Left) Transfected cells expressing full-length p10-2HAN were pulse-labeled and chased for the indicated times in minutes. Radiolabeled cells were solubilized in RIPA buffer or double-strength RIPA buffer, and p10 was immunoprecipitated with anti-p10 antiserum (α p10), resolved by SDS-PAGE, and visualized by fluorography. (Right) Same analysis with a control protein, the soluble sigma C protein of avian reovirus. (B) Full-length p10-2HAN (p10), p10ecto− (ecto−), or p10e, with two N-terminal HA epitope tags (HA) and the cleavable HA signal peptide (SP), were expressed in transfected cells. The stability of the various p10 constructs was assessed by pulse-chase analysis as described for panel A. The nature of the various constructs is diagrammed at the bottom.
FIG. 2.
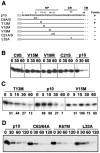
Residues in the p10 HP influence p10 degradation. (A) Sequence and schematic representation of the p10 ectodomain. Numbers refer to amino acid residues. The locations of the HP, the CR in the ARV and NBV p10 proteins, and the start of the downstream TM are indicated. The locations of various site-specific substitutions in the ARV p10-2HAN construct are indicated. Constructs are named by using the single-letter amino acid code to indicate the identity of the authentic amino acid, its position, and the identity of the substitution. A/S indicates a substitution with either Ala or Ser. (B) Degradation rates for authentic p10-2HAN (p10) and constructs containing single amino acid substitutions, assessed by pulse-chase analysis (chase times are indicated in minutes) as described in the legend to Fig. 1. (C) Time course analysis of p10 degradation performed as described for panel B but with more precise time points (in minutes) to compare authentic p10-2HAN (p10) to two p10 constructs containing single residue substitutions; one of these constructs retains fusion activity (T13M), and one is nonfusogenic (V15M). Numbers below the gel lanes indicate the percentages of p10 detected after various chase times relative to that in the pulse-labeled sample with no chase, as determined by image analysis of the fluorogram. (D) Degradation rates for p10-2HAN (p10) and constructs containing single amino acid substitutions in the endodomain dicysteine (C63/64A) or polybasic (K67M) motifs or in the ectodomain conserved region (L32A), assessed as described for panel B.
FIG. 3.
p10 localizes to the ER. (Left) Cells transfected with a p10 expression plasmid were fixed and permeabilized, and the subcellular distribution of p10 was examined by confocal microscopy with anti-p10 polyclonal antiserum (α p10) and FITC-conjugated secondary antibody. (Middle) A similar analysis was performed with anticalnexin antibody (α calnexin) and Texas red-conjugated secondary antibody to reveal the distribution of an ER-localized protein. (Right) Merge of the p10 and calnexin panels, showing extensive colocalization of the two proteins. Scale bars, 10 μm.
FIG. 4.
Degradation of p10 occurs following membrane insertion and is altered by a proteasome inhibitor. (A) Pulse-chase analysis for the indicated times (in minutes) was performed with p10-transfected cells as described in the legend to Fig. 1. After the pulse or each chase, the membrane fraction was isolated, and the presence of membrane-associated p10 was determined by immunoprecipitation, SDS-PAGE, and fluorography. (B) Pulse-chase analysis for the indicated times (in minutes) was performed with p10-transfected cells as described in the legend to Fig. 1, except that cells were incubated during the pulse-chase in the presence of inhibitors of lysosomal proteases (E64C) or lysosome acidification (chloroquine [chlor] or methylamine [MA]) or a proteasome inhibitor (MG132).
FIG. 5.
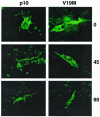
Surface-localized p10 is relatively stable. The stabilities of surface-localized p10-2HAN (p10) and the V19M construct were assessed by surface staining of live transfected cells with anti-HA monoclonal antibody. Following antibody addition, the presence of surface-bound antibody specifically attached to the HA-tagged, surface-localized p10 ectodomain was monitored over time (indicated in minutes on the right). Fluorescence images were captured under identical parameters for comparison of the intensities of fluorescence.
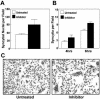
FIG. 6.
Inhibition of proteasome activity leads to enhanced p10-mediated syncytium formation. Cell monolayers were transfected with a p10 expression plasmid. At 15 h posttransfection, as syncytia were beginning to appear, the culture medium was removed and replaced with medium lacking or containing the proteasome inhibitor MG132. Treated monolayers were incubated for a further 4 to 6 h prior to Giemsa staining to reveal syncytial foci. Microscopic examination was used to quantify the average number of syncytial nuclei per field at 6 h posttreatment (A) or the average number of syncytia per field at 4 and 6 h posttreatment (B). Results are presented as the mean and standard deviation of triplicate samples from a representative experiment. The numbers and sizes of the syncytia present in Giemsa-stained monolayers at 6 h posttreatment were captured by light microscopy at a magnification of ×194 (C).
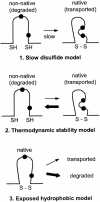
FIG. 7.
Models of p10 degradation. Three possible models to explain the influence of the p10 ectodomain and the HP on p10 degradation by the ERAD pathway are shown. The first model implies that the slow formation of an intramolecular disulfide bond (S—S) leads to exposed thiol (SH) or hydrophobic residues (filled circles) in the nonnative structure which target p10 for degradation. The second model implies that it is not the rate of folding of the p10 ectodomain but rather a low thermodynamic stability of the native structure that leads to transient and repeated exposure of thiol or hydrophobic residues. The third model implies that hydrophobic residues are naturally surface exposed in the native p10 ectodomain structure, leading to recognition and degradation of the majority of p10.
Similar articles
- Unusual topological arrangement of structural motifs in the baboon reovirus fusion-associated small transmembrane protein.
Dawe S, Corcoran JA, Clancy EK, Salsman J, Duncan R. Dawe S, et al. J Virol. 2005 May;79(10):6216-26. doi: 10.1128/JVI.79.10.6216-6226.2005. J Virol. 2005. PMID: 15858006 Free PMC article. - Structural and functional properties of an unusual internal fusion peptide in a nonenveloped virus membrane fusion protein.
Shmulevitz M, Epand RF, Epand RM, Duncan R. Shmulevitz M, et al. J Virol. 2004 Mar;78(6):2808-18. doi: 10.1128/jvi.78.6.2808-2818.2004. J Virol. 2004. PMID: 14990700 Free PMC article. - Avian reovirus: structure and biology.
Benavente J, Martínez-Costas J. Benavente J, et al. Virus Res. 2007 Feb;123(2):105-19. doi: 10.1016/j.virusres.2006.09.005. Epub 2006 Oct 2. Virus Res. 2007. PMID: 17018239 Review. - Reovirus FAST proteins: virus-encoded cellular fusogens.
Ciechonska M, Duncan R. Ciechonska M, et al. Trends Microbiol. 2014 Dec;22(12):715-24. doi: 10.1016/j.tim.2014.08.005. Epub 2014 Sep 19. Trends Microbiol. 2014. PMID: 25245455 Review.
Cited by
- Characterization of a New Toti-like Virus in Sea Bass, Dicentrarchus labrax.
Louboutin L, Cabon J, Beven V, Hirchaud E, Blanchard Y, Morin T. Louboutin L, et al. Viruses. 2023 Dec 13;15(12):2423. doi: 10.3390/v15122423. Viruses. 2023. PMID: 38140664 Free PMC article. - A compact, multifunctional fusion module directs cholesterol-dependent homomultimerization and syncytiogenic efficiency of reovirus p10 FAST proteins.
Key T, Duncan R. Key T, et al. PLoS Pathog. 2014 Mar 20;10(3):e1004023. doi: 10.1371/journal.ppat.1004023. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24651689 Free PMC article. - A virus-encoded cell-cell fusion machine dependent on surrogate adhesins.
Salsman J, Top D, Barry C, Duncan R. Salsman J, et al. PLoS Pathog. 2008 Mar 7;4(3):e1000016. doi: 10.1371/journal.ppat.1000016. PLoS Pathog. 2008. PMID: 18369467 Free PMC article. - Liposome reconstitution of a minimal protein-mediated membrane fusion machine.
Top D, de Antueno R, Salsman J, Corcoran J, Mader J, Hoskin D, Touhami A, Jericho MH, Duncan R. Top D, et al. EMBO J. 2005 Sep 7;24(17):2980-8. doi: 10.1038/sj.emboj.7600767. Epub 2005 Aug 4. EMBO J. 2005. PMID: 16079913 Free PMC article. - Novel strand-specific qPCR assay elucidates differences in viral replication kinetics between different strains of avian reovirus.
Harrell TL, Alvarez-Narvaez S, Read QD, Conrad SJ. Harrell TL, et al. Microbiol Spectr. 2025 Jun 3;13(6):e0314024. doi: 10.1128/spectrum.03140-24. Epub 2025 Apr 30. Microbiol Spectr. 2025. PMID: 40304489 Free PMC article.
References
- Beal, R. E., D. Toscano-Cantaffa, P. Young, M. Rechsteiner, and C. M. Pickart. 1998. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry 37:2925-2934. - PubMed
- Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2002. Modification of late membrane permeability in avian reovirus-infected cells. J. Biol. Chem. 277:17789-17796. - PubMed
- Bohley, P. 1996. Surface hydrophobicity and intracellular degradation of proteins. Biol. Chem. 377:425-435. - PubMed
- Chen, Y. A., and R. H. Scheller. 2001. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell. Biol. 2:98-106. - PubMed
- Ciechanover, A., and A. L. Schwartz. 1994. The ubiquitin-mediated proteolytic pathway: mechanisms of recognition of the proteolytic substrate and involvement in the degradation of native cellular proteins. FASEB J. 8:182-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources