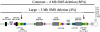Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates - PubMed (original) (raw)
Comparative Study
doi: 10.1086/422016. Epub 2004 May 13.
Affiliations
- PMID: 15148657
- PMCID: PMC1182010
- DOI: 10.1086/422016
Comparative Study
Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates
Christine J Shaw et al. Am J Hum Genet. 2004 Jul.
Abstract
Several homologous recombination "hotspots," or sites of positional preference for strand exchanges, associated with recurrent deletions and duplications have been reported within large low-copy repeats (LCRs). Recently, such a hotspot was identified in patients with the Smith-Magenis syndrome (SMS) common deletion of approximately 4 Mb or a reciprocal duplication within the KER gene cluster of the SMS-REP LCRs, in which 50% of analyzed strand exchanges resulting in deletion and 23% of those resulting in duplication occurred. Here, we report an additional recombination hotspot within LCR17pA and LCR17pD, which serve as alternative substrates for nonallelic homologous recombination that results in large (approximately 5 Mb) deletions of 17p11.2, which include the SMS region. Using polymerase-chain-reaction mapping of somatic cell hybrid lines, we refined the breakpoints of six deletions within these LCRs. Sequence analysis of the recombinant junctions revealed that all six strand exchanges occurred within a 524-bp interval, and four of them occurred within an AluSq/x element. This interval represents only 0.5% of the 124-kb stretch of 98.6% sequence identity between LCR17pA and LCR17pD. A search for potentially stimulating sequence motifs revealed short AT-rich segments flanking the recombination hotspot. Our findings indicate that alternative LCRs can mediate rearrangements, resulting in haploinsufficiency of the SMS critical region, and reimplicate homologous recombination as a major mechanism for genomic disorders.
Figures
Figure 1
Comparison of common and uncommon deletions of SMS. Proximal chromosome 17p is depicted at the bottom, with the position of LCRs shown. The common deletion is shown above, and the large deletions of SMS (discussed in the main text) are shown below. The solid horizontal lines represent the undeleted chromosome segments, and the dotted lines represent the deleted chromosome segments. The numbers identifying analyzed patients with large deletions are listed (right). The arrows indicate the distal and proximal breakpoints of the large deletions within LCR17pA and LCR17pD, respectively.
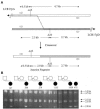
Figure 2
Detection of a patient-specific junction fragment. A, Schematic representation of the breakpoint regions in LCR17pA (white rectangle) and LCR17pD (gray rectangle). The dotted line represents the proposed crossover event between the LCRs; the 2.0-kb junction fragment results from this crossover. Small unblackened arrows denote the primers used for amplification of the 5.2-kb product containing the breakpoint region. _Acl_I restriction sites and expected fragment sizes upon digestion of the PCR product are shown (not drawn to scale). B, Results of assay. The sizes of the bands are shown to the right; the patients and their parents are represented by the pedigrees at the top. All six patients, but none of the parents, have the expected 2.0-kb junction fragment, indicating that the six strand exchanges occurred within this interval. The 5.2-kb undigested fragment is visible and presumably a result of incomplete digestion. The 0.5-kb fragment from LCR17pA is not shown.
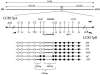
Figure 3
Fine mapping of the deletion breakpoints within the 2.0-kb _Acl_I junction fragment. At the top is a diagram of the position of the hotspot (dotted box) within the LCR sequence, and a finer-scale view of the hotspot itself is shown with the telomere (tel) on the left and the centromere (cen) on the right. Paralogous sequence variants, or nucleotide _cis_-morphisms, between LCR17pA (above the line) and LCR17pD (below the line) are represented by uppercase letters. Polymorphic nucleotides are represented by lowercase letters. Restriction enzyme consensus sequence _cis_-morphisms (including the _Acl_I sites used to isolate a patient-specific junction fragment) are also shown. Alu elements are depicted by the diagonally striped rectangles, and AT-rich segments are represented by the blackened boxes labeled A, B, or C (C represents the AT-rich segment distal to the hotspot, found only in LCR17pD). In the lower portion of the figure, patient numbers are given (right). Each circle represents a _cis_-morphic nucleotide within the patient’s recombinant LCR17pA/D. Unblackened circles denote nucleotides matching LCR17pA, and blackened circles denote nucleotides matching LCR17pD. Bold lines are drawn between the two _cis_-morphisms, in which the transition from distal-like to proximal-like sequence occurs for each patient. The sizes of these intervals are given at the bottom (not drawn to scale).
Similar articles
- Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2.
Bi W, Park SS, Shaw CJ, Withers MA, Patel PI, Lupski JR. Bi W, et al. Am J Hum Genet. 2003 Dec;73(6):1302-15. doi: 10.1086/379979. Epub 2003 Nov 24. Am J Hum Genet. 2003. PMID: 14639526 Free PMC article. - Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs.
Park SS, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR. Park SS, et al. Genome Res. 2002 May;12(5):729-38. doi: 10.1101/gr.82802. Genome Res. 2002. PMID: 11997339 Free PMC article. - Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2.
Shaw CJ, Bi W, Lupski JR. Shaw CJ, et al. Am J Hum Genet. 2002 Nov;71(5):1072-81. doi: 10.1086/344346. Epub 2002 Oct 9. Am J Hum Genet. 2002. PMID: 12375235 Free PMC article. - Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications.
Yatsenko SA, Treadwell-Deering D, Krull K, Lewis RA, Glaze D, Stankiewicz P, Lupski JR, Potocki L. Yatsenko SA, et al. Am J Med Genet A. 2005 Oct 1;138A(2):175-80. doi: 10.1002/ajmg.a.30948. Am J Med Genet A. 2005. PMID: 16152635 Review. - New developments in Smith-Magenis syndrome (del 17p11.2).
Gropman AL, Elsea S, Duncan WC Jr, Smith AC. Gropman AL, et al. Curr Opin Neurol. 2007 Apr;20(2):125-34. doi: 10.1097/WCO.0b013e3280895dba. Curr Opin Neurol. 2007. PMID: 17351481 Review.
Cited by
- Case Report: Balanced Reciprocal Translocation t (17; 22) (p11.2; q11.2) and 10q23.31 Microduplication in an Infertile Male Patient Suffering From Teratozoospermia.
Huang S, Wu H, Qi Y, Wei L, Lv X, He Y. Huang S, et al. Front Genet. 2022 May 26;13:797813. doi: 10.3389/fgene.2022.797813. eCollection 2022. Front Genet. 2022. PMID: 35719406 Free PMC article. - Genomic regions associated with microdeletion/microduplication syndromes exhibit extreme diversity of structural variation.
Mostovoy Y, Yilmaz F, Chow SK, Chu C, Lin C, Geiger EA, Meeks NJL, Chatfield KC, Coughlin CR, Surti U, Kwok PY, Shaikh TH. Mostovoy Y, et al. Genetics. 2021 Feb 9;217(2):iyaa038. doi: 10.1093/genetics/iyaa038. Genetics. 2021. PMID: 33724415 Free PMC article. - Biogeochemical behavior of nickel under different abiotic stresses: toxicity and detoxification mechanisms in plants.
Ameen N, Amjad M, Murtaza B, Abbas G, Shahid M, Imran M, Naeem MA, Niazi NK. Ameen N, et al. Environ Sci Pollut Res Int. 2019 Apr;26(11):10496-10514. doi: 10.1007/s11356-019-04540-4. Epub 2019 Mar 5. Environ Sci Pollut Res Int. 2019. PMID: 30835069 Review. - Identification of a RAI1-associated disease network through integration of exome sequencing, transcriptomics, and 3D genomics.
Loviglio MN, Beck CR, White JJ, Leleu M, Harel T, Guex N, Niknejad A, Bi W, Chen ES, Crespo I, Yan J, Charng WL, Gu S, Fang P, Coban-Akdemir Z, Shaw CA, Jhangiani SN, Muzny DM, Gibbs RA, Rougemont J, Xenarios I, Lupski JR, Reymond A. Loviglio MN, et al. Genome Med. 2016 Nov 1;8(1):105. doi: 10.1186/s13073-016-0359-z. Genome Med. 2016. PMID: 27799067 Free PMC article. - Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over.
Liu P, Lacaria M, Zhang F, Withers M, Hastings PJ, Lupski JR. Liu P, et al. Am J Hum Genet. 2011 Oct 7;89(4):580-8. doi: 10.1016/j.ajhg.2011.09.009. Am J Hum Genet. 2011. PMID: 21981782 Free PMC article.
References
Electronic-Database Information
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
References
- Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, Inoue K, Lupski JR (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–72810.1101/gr.73702 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous