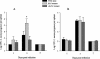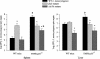Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence - PubMed (original) (raw)
Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence
Lone Dons et al. Infect Immun. 2004 Jun.
Abstract
The flagellum protein flagellin of Listeria monocytogenes is encoded by the flaA gene. Immediately downstream of flaA, two genes, cheY and cheA, encoding products with homology to chemotaxis proteins of other bacteria, are located. In this study we constructed deletion mutants with mutations in flaA. cheY, and cheA to elucidate their role in the biology of infection with L. monocytogenes. The DeltacheY, DeltacheA, and double-mutant DeltacheYA mutants, but not DeltaflaA mutant, were motile in liquid media. However, the DeltacheA mutant had impaired swarming and the DeltacheY and DeltacheYA mutants were unable to swarm on soft agar plates, suggesting that cheY and cheA genes encode proteins involved in chemotaxis. The DeltaflaA, DeltacheY, DeltacheA, and DeltacheYA mutants (grown at 24 degrees C) showed reduced association with and invasion of Caco-2 cells compared to the wild-type strain. However, spleens from intragastrically infected BALB/c and C57BL/6 mice showed larger and similar numbers of the DeltaflaA and DeltacheYA mutants, respectively, compared to the wild-type controls. Such a discrepancy could be explained by the fact that tumor necrosis factor receptor p55 deficient mice showed dramatically exacerbated susceptibility to the wild-type but unchanged or only slightly increased levels of the DeltaflaA or DeltacheYA mutant. In summary, we show that listerial flaA. cheY, and cheA gene products facilitate the initial contact with epithelial cells and contribute to effective invasion but that flaA could also be involved in the triggering of immune responses.
Figures
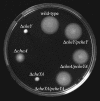
FIG. 1.
Swarming of L. monocytogenes mutants in semisolid agar. The L. monocytogenes WT strain 12067 and the Δ_flaA_, Δ_cheY_, Δ_cheA_, and Δ_cheYA_ mutants were stabbed into semisolid agar plates (tryptic soy broth plus 0.25% agar). The plates were incubated at 24°C (A) or 37°C (B) for 24 h.
FIG. 2.
Western blot analysis of whole-cell lysates of the Δ_flaA_, Δ_cheY_, Δ_cheA_, and Δ_cheYA_ mutants grown at 24°C (5 × 107 CFU). Samples were run on a 12.5% polyacrylamide gel and incubated with a monoclonal antibody specific for the L. monocytogenes 4b flagellin. Lanes: 1, L. monocytogenes 12067 (WT); 2, Δ_flaA_ mutant; 3, Δ_cheY_ mutant; 4, Δ_cheA_ mutant; 5, Δ_cheYA_ mutant. Molecular mass standards are shown on the left.
FIG. 3.
Electron micrographs of the flagellated L. monocytogenes WT strain (A), the nonflagellated Δ_flaA_ mutant (B), the Δ_cheY_ mutant (C), the Δ_cheA_ mutant (D), and the Δ_cheYA_ mutant (E). Bacteria were grown at 24°C to late logarithmic growth phase, applied to carbon-coated grids, shadowed with 2% uranyl acetate, and examined under a transmission electron microscope. The scale bar, shown in panel C and valid for all panels, represents 1.5 μm (B), 1 μm (A, C, and E), and 0.5 μm (D).
FIG. 4.
Swarming of complemented Δ_cheY_, Δ_cheA_, and Δ_cheYA_ mutants in semisolid agar. The Δ_cheY_, Δ_cheA_, and Δ_cheYA_ mutants with plasmids carrying the cheY (p_cheY_) or cheY/cheA (p_cheYA_) genes were stabbed into a semisolid agar plate. The WT strain and the Δ_cheY_, Δ_cheA_ and Δ_cheYA_ mutants carrying the parent vector pAT19 served as controls. The plate was incubated at 24°C for 30 h.
FIG. 5.
CFU in the spleens of i.g. infected BALB/c mice. L. monocytogenes WT and the Δ_flaA_ and Δ_cheYA_ mutants were grown at either 24°C (A) or 37°C (B) before being used to infect BALB/c mice (five mice per group). The mice were sacrified at the indicated time points after infection, and their spleens were homogenized and plated. The means and standard errors of the mean are shown. An asterisk indicates a significant difference in the log CFU of the mutant compared to the WT (P < 0.05 as determined by Student's t test). A representative of two independent experiments is shown.
FIG. 6.
CFU after i.g. infection of TNFR-p55−/− and WT mice. L. monocytogenes WT and mutant strains were inoculated i.g. into WT or TNFR-p55−/− mice (six mice per group). Three days after infection, the mice were sacrified and bacterial loads (CFU) in their spleens and livers were measured. The means and standard errors of the mean are shown. The asterisk indicates a significant difference in the log CFU of a bacterial mutant relative to the WT (P < 0.05 as determined by Student's t test). The pound sign indicates a significant difference between two groups of mice infected with the same bacterial strain (P < 0.05 as determined by Student's t test).
Similar articles
- Cold-Shock Domain Family Proteins (Csps) Are Involved in Regulation of Virulence, Cellular Aggregation, and Flagella-Based Motility in Listeria monocytogenes.
Eshwar AK, Guldimann C, Oevermann A, Tasara T. Eshwar AK, et al. Front Cell Infect Microbiol. 2017 Oct 26;7:453. doi: 10.3389/fcimb.2017.00453. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29124040 Free PMC article. - Role of FliF and FliI of Listeria monocytogenes in flagellar assembly and pathogenicity.
Bigot A, Pagniez H, Botton E, Fréhel C, Dubail I, Jacquet C, Charbit A, Raynaud C. Bigot A, et al. Infect Immun. 2005 Sep;73(9):5530-9. doi: 10.1128/IAI.73.9.5530-5539.2005. Infect Immun. 2005. PMID: 16113269 Free PMC article. - Response regulation in bacterial chemotaxis.
Lukat GS, Stock JB. Lukat GS, et al. J Cell Biochem. 1993 Jan;51(1):41-6. doi: 10.1002/jcb.240510109. J Cell Biochem. 1993. PMID: 8381790 Review.
Cited by
- Deciphering the impact of exogenous fatty acids on Listeria monocytogenes at low temperature by transcriptome analysis.
Quilleré A, Darsonval M, Papadochristopoulos A, Amoros A, Nicolas P, Dubois-Brissonnet F. Quilleré A, et al. Front Microbiol. 2024 Sep 4;15:1441784. doi: 10.3389/fmicb.2024.1441784. eCollection 2024. Front Microbiol. 2024. PMID: 39328916 Free PMC article. - Going against the grain: chemotaxis and infection in Vibrio cholerae.
Butler SM, Camilli A. Butler SM, et al. Nat Rev Microbiol. 2005 Aug;3(8):611-20. doi: 10.1038/nrmicro1207. Nat Rev Microbiol. 2005. PMID: 16012515 Free PMC article. Review. - The Listeria monocytogenes Bile Stimulon under Acidic Conditions Is Characterized by Strain-Specific Patterns and the Upregulation of Motility, Cell Wall Modification Functions, and the PrfA Regulon.
Guariglia-Oropeza V, Orsi RH, Guldimann C, Wiedmann M, Boor KJ. Guariglia-Oropeza V, et al. Front Microbiol. 2018 Feb 6;9:120. doi: 10.3389/fmicb.2018.00120. eCollection 2018. Front Microbiol. 2018. PMID: 29467736 Free PMC article. - Transcriptome analysis of Listeria monocytogenes exposed to biocide stress reveals a multi-system response involving cell wall synthesis, sugar uptake, and motility.
Casey A, Fox EM, Schmitz-Esser S, Coffey A, McAuliffe O, Jordan K. Casey A, et al. Front Microbiol. 2014 Feb 28;5:68. doi: 10.3389/fmicb.2014.00068. eCollection 2014. Front Microbiol. 2014. PMID: 24616718 Free PMC article. - Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes.
Orsi RH, Sun Q, Wiedmann M. Orsi RH, et al. BMC Evol Biol. 2008 Aug 12;8:233. doi: 10.1186/1471-2148-8-233. BMC Evol Biol. 2008. PMID: 18700032 Free PMC article.
References
- Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. - PubMed
- Bischoff, D. S., R. B. Bourret, M. L. Kirsch, and G. W. Ordal. 1993. Purification and characterization of Bacillus subtilis CheY. Biochemistry 32:9256-9261. - PubMed
- Bischoff, D. S., and G. W. Ordal. 1991. Sequence and characterization of Bacillus subtilis CheB, a homolog of Escherichia coli CheY, and its role in a different mechanism of chemotaxis. J. Biol. Chem. 266:12301-12305. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources