Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy - PubMed (original) (raw)
Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy
Per E Strømhaug et al. Mol Biol Cell. 2004 Aug.
Abstract
Delivery of proteins and organelles to the vacuole by autophagy and the cytoplasm to vacuole targeting (Cvt) pathway involves novel rearrangements of membrane resulting in the formation of vesicles that fuse with the vacuole. The mechanism of vesicle formation and the origin of the membrane are complex issues still to be resolved. Atg18 and Atg21 are proteins essential to vesicle formation and together with Ygr223c form a novel family of phosphoinositide binding proteins that are associated with the vacuole and perivacuolar structures. Their localization requires the activity of Vps34, suggesting that phosphatidylinositol(3)phosphate may be essential for their function. The activity of Atg18 is vital for all forms of autophagy, whereas Atg21 is required for the Cvt pathway but not for nitrogen starvation-induced autophagy. The loss of Atg21 results in the absence of Atg8 from the pre-autophagosomal structure (PAS), which may be ascribed to a reduced rate of conjugation of Atg8 to phosphatidylethanolamine. A similar defect in localization of a second ubiquitin-like conjugate, Atg12-Atg5, suggests that Atg21 may be involved in the recruitment of membrane to the PAS.
Figures
Figure 1.
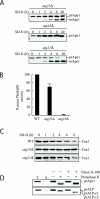
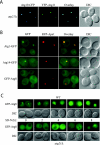
Atg21 is required for the Cvt pathway. (A) _atg21_Δ cells were grown in YPD and shifted to SD-N to induce autophagy. The _atg21_Δ cells mature prApe1 under starvation conditions more rapidly than _atg11_Δ, whereas no maturation is seen in _atg18_Δ cells. (B) Pho8Δ60, a marker for nonspecific autophagy, is delivered to the vacuole of _atg21_Δ cells in nitrogen starvation conditions. WT (TN124), _atg18_Δ (JGY20), and _atg21_Δ (PSY107) cells were grown as described in A. Cells were shifted to SD-N, samples collected, and protein extracts assayed for alkaline phosphatase activity. The activity in the wild-type sample was set to 100% and the other activities normalized relative to wild-type. The _atg21_Δ results represent the mean and S.E. of three experiments, whereas the _atg18_Δ results are based on duplicate samples from the same experiment. (C) _atg21_Δ and _ygr223c_Δ cells are not deficient in peroxisome degradation. Cells were shifted from oleic acid-containing medium to SD-N, and pexophagy was monitored by Western blot by using an antibody against peroxisomal thiolase (Fox3). The result for the _ygr223c_Δ strain was essentially the same as that shown for _atg21_Δ. (D) Atg21 is required for Cvt vesicle formation. _atg21_Δ _pep4_Δ (PSY2) cells were converted to spheroplasts and subjected to a protease protection assay performed as described in MATERIALS AND METHODS.
Figure 2.
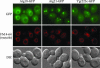
Atg21 is associated with the vacuole and prevacuolar structures. Wild-type (SEY6210) cells were chromosomally tagged with GFP at the ATG18 (PSY62), ATG21 (PSY63), and YGR223c (PSY100) loci, respectively, and stained with FM 4-64 to label the vacuoles. Cells were grown to early mid-log phase in YPD before viewing. DIC, differential interference contrast.
Figure 3.
Atg18, Atg21, and Ygr223c localization is dependent on PtdIns 3-kinase activity. Strains PSY291, PSY292, PSY293, PSY62, PSY63, PSY100, PSY146, and PSY290 expressing Atg18-GFP, Atg21-GFP, or Ygr223c-GFP from the respective chromosomal loci in the vps34ts and wild-type SEY6210 (A) or fab1-2ts background (B) were grown at 26°C to early mid-log phase in SMD and shifted to 38°C for 10 (A) or 40 min (B). Cells were viewed by fluorescence microscopy as described in the legend to Figure 2. Loss of PtdIns 3-kinase but not PtdIns(3)P 5-kinase activity at the nonpermissive temperature resulted in loss of localization of Atg18 and Atg21. DIC, differential interference contrast.
Figure 4.
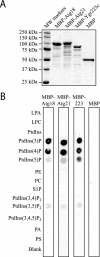
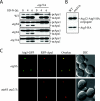
Atg21 binds to phosphoinositides in vitro. (A) MBP, MBP-Atg18, MBP-Atg21, and MBP-Ygr223c were purified from E. coli as described in MATERIALS AND METHODS. Protein (1 μg) was loaded on a gel, and the gel was stained with Coomassie Brilliant Blue G. (B) PIP Strips (Echelon) containing lipids immobilized on nitrocellulose were incubated with 100 ng/ml MBP, MBP-Atg18, MBP-Atg21, or MBP-Ygr223c as described in MATERIALS AND METHODS.
Figure 5.
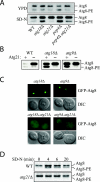
Arginine343 and arginine344 are required for function of Atg21. (A) _atg21_Δ cells (PSY7) expressing GFP-Atg21 (pPS123) but not GFP-Atg21R343,344K (pPS125) mature prApe1. (B) GFP-Atg21R343,344K does not localize to the vacuole and punctate structures. DIC, differential interference contrast.
Figure 6.
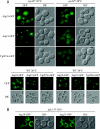
Localization of Atg proteins to the PAS in _atg21_Δ cells. (A) Atg19-CFP and Atg11-YFP colocalize in _atg21_Δ cells. PSY6 (_atg21_Δ) cells were transformed with plasmids expressing Atg19-CFP (pCVT19CFP(414)) and YFP-Atg11 (pPS97), grown in SMD to mid-log phase and analyzed by fluorescent microscopy. (B) GFP-Atg8 is not localized to the PAS in _atg21_Δ cells. _atg21_Δ cells expressing Atg1-GFP (PSY227) or Atg14-GFP (PSY228) from the chromosomal loci, and also expressing RFP-prApe1 from the endogenous promoter on an integrated plasmid, or _atg21_Δ _atg8_Δ cells expressing GFP-Atg8 (PSY226) behind the endogenous promoter from an integrated plasmid, and also expressing RFP-prApe1 from the endogenous promoter on a centromeric plasmid, were grown in YPD to mid-log phase before viewing. Localization of Atg2-GFP was essentially the same as that shown for Atg1-GFP. (C) Transport of GFP-Atg8 to the vacuole is reduced in _atg21_Δ cells. Wild-type (WT, SEY6210) and _atg21_Δ cells (PSY7) were transformed with a plasmid expressing GFP-Atg8 from the _CUP1-_promoter (pCuGFP-AUT7(416)), grown to mid-log phase in SMD before the addition of 1 mM phenylmethylsulfonyl fluoride and starved for nitrogen for up to 8 h as indicated. DIC, differential interference contrast.
Figure 7.
Lipidation of Atg8 is reduced in _atg21_Δ cells. (A) Atg8 lipidation in rich medium versus nitrogen starvation medium. Cells were grown in YPD to early mid-log phase and then starved for 3 h in SD-N. Atg8-PE was separated from Atg8 by 12% SDS page gels containing 6% urea (Kirisako et al., 2000). All strains are of BY4742 background. (B) Deletion of ATG21 antagonizes the accumulation of Atg8-PE in autophagy mutants. WT (SEY6210), _atg21_Δ (PSY7), _atg18_Δ (JGY3), _atg18_Δ _atg21_Δ (PSY167), _atg9_Δ (JKY007), and _atg9_Δ _atg21_Δ (PSY191) cells were starved for 4 h in SD-N, and Atg8-PE was analyzed by Western blot. (C) Deletion of ATG21 antagonizes the accumulation of GFP-Atg8 at the PAS in autophagy mutants. Strains used in B containing a plasmid expressing GFP-Atg8 from the CUP1 promoter were grown to mid-log phase in SMD and then starved for 4 h in SD-N. (D) _atg21_Δ cells have a reduced rate of Atg8 lipidation. Cells of BY4742 background were labeled and Atg8 immunoprecipitated as described in MATERIALS AND METHODS and analyzed by 12% SDS-urea gels.
Figure 8.
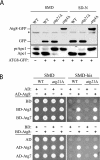
The conjugation system responsible for posttranslational modification of Atg8 is functional in the _atg21_Δ mutant. (A) Atg4 is able to recognize Atg8 and carry out proteolytic cleavage at the C terminus. A plasmid encoding Atg8-GFP was transformed into the wild-type (SEY6210), _atg4_Δ (WPHYD2) or _atg21_Δ (PSY5) strain. Cells were grown in SMD to early log phase (A600 = 0.6) and then either retained in SMD or nitrogen starved in SD-N medium for 3 h. Protein extracts were analyzed by Western blot by using antiserum to Ape1 or anti-GFP monoclonal antibodies. The asterisk denotes a contaminating band that cross-reacts with the anti-GFP antibody. (B) Atg7 E1-like and Atg3 E2 enzymes are able to activate and conjugate Atg8. A yeast two-hybrid analysis was carried out as described in MATERIALS AND METHODS.
Figure 9.
Atg21 is required for Atg5-GFP accumulation at the PAS. (A) Cells lacking both Atg8 and Atg21 do not mature prApe1 upon starvation. WT (SEY6210), single mutants (WPHYD7, WPHYD2, AHY001, and PSY7) and double mutants (PSY139, PSY128, and PSY190) were grown in YPD to mid-log phase and starved for 4 and 6 h before being analyzed by Western blot by using an antibody against Ape1. (B) Atg21 is not required for conjugation of Atg12 to Atg5. WT (PSY66) and _atg21_Δ (PSY155) cells expressing Atg5-HA from the chromosomal locus were grown to early mid-log phase in YPD and subjected to Western blot by using an antibody against HA. (C) Atg21 is required for Atg5-GFP accumulation at the PAS. _atg8_Δ (PSY285) and _atg8_Δ _atg21_Δ (PSY286) cells expressing Atg5-GFP from the chromosomal locus and RFP-prApe1 from an integrating plasmid were grown to early mid-log phase in YPD before being viewed by fluorescence microscopy as described in the legend to Figure 6. DIC, differential interference contrast.
Similar articles
- Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy.
Nair U, Cao Y, Xie Z, Klionsky DJ. Nair U, et al. J Biol Chem. 2010 Apr 9;285(15):11476-88. doi: 10.1074/jbc.M109.080374. Epub 2010 Feb 12. J Biol Chem. 2010. PMID: 20154084 Free PMC article. - PI3P binding by Atg21 organises Atg8 lipidation.
Juris L, Montino M, Rube P, Schlotterhose P, Thumm M, Krick R. Juris L, et al. EMBO J. 2015 Apr 1;34(7):955-73. doi: 10.15252/embj.201488957. Epub 2015 Feb 17. EMBO J. 2015. PMID: 25691244 Free PMC article. - Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway.
Meiling-Wesse K, Barth H, Voss C, Eskelinen EL, Epple UD, Thumm M. Meiling-Wesse K, et al. J Biol Chem. 2004 Sep 3;279(36):37741-50. doi: 10.1074/jbc.M401066200. Epub 2004 Jun 11. J Biol Chem. 2004. PMID: 15194695 - Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation.
Teter SA, Klionsky DJ. Teter SA, et al. Semin Cell Dev Biol. 2000 Jun;11(3):173-9. doi: 10.1006/scdb.2000.0163. Semin Cell Dev Biol. 2000. PMID: 10906274 Review. - [Cytoplasm to vacuole targeting pathway in yeast].
Shintani T. Shintani T. Tanpakushitsu Kakusan Koso. 2006 Aug;51(10 Suppl):1480-3. Tanpakushitsu Kakusan Koso. 2006. PMID: 16922423 Review. Japanese. No abstract available.
Cited by
- Autophagy - An Emerging Anti-Aging Mechanism.
Gelino S, Hansen M. Gelino S, et al. J Clin Exp Pathol. 2012 Jul 12;Suppl 4:006. doi: 10.4172/2161-0681.s4-006. J Clin Exp Pathol. 2012. PMID: 23750326 Free PMC article. - Qualitative and quantitative characterization of protein-phosphoinositide interactions with liposome-based methods.
Busse RA, Scacioc A, Hernandez JM, Krick R, Stephan M, Janshoff A, Thumm M, Kühnel K. Busse RA, et al. Autophagy. 2013 May;9(5):770-7. doi: 10.4161/auto.23978. Epub 2013 Feb 27. Autophagy. 2013. PMID: 23445924 Free PMC article. - Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy.
Zhao L, You W, Sun D, Xu H, You X, Xu H, Wu Z, Xie Z, Liang Y. Zhao L, et al. Int J Mol Sci. 2022 Aug 23;23(17):9550. doi: 10.3390/ijms23179550. Int J Mol Sci. 2022. PMID: 36076954 Free PMC article. - The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy.
Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. Yen WL, et al. J Cell Biol. 2010 Jan 11;188(1):101-14. doi: 10.1083/jcb.200904075. J Cell Biol. 2010. PMID: 20065092 Free PMC article. - Autophagosome formation in mammalian cells.
Burman C, Ktistakis NT. Burman C, et al. Semin Immunopathol. 2010 Dec;32(4):397-413. doi: 10.1007/s00281-010-0222-z. Epub 2010 Aug 26. Semin Immunopathol. 2010. PMID: 20740284 Review.
References
- Barth, H., Meiling-Wesse, K., Epple, U.D., and Thumm, M. (2001). Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 508, 23-28. - PubMed
- Barth, H., Meiling-Wesse, K., Epple, U.D., and Thumm, M. (2002). Mai1p is essential for maturation of proaminopeptidase I but not for autophagy. FEBS Lett. 512, 173-179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases