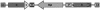CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis - PubMed (original) (raw)
. 2004 Jun;16(6):1633-43.
doi: 10.1105/tpc.021378. Epub 2004 May 21.
Affiliations
- PMID: 15155891
- PMCID: PMC490051
- DOI: 10.1105/tpc.021378
CENTRIN2 modulates homologous recombination and nucleotide excision repair in Arabidopsis
Jean Molinier et al. Plant Cell. 2004 Jun.
Abstract
A genetic screen of a population of Arabidopsis thaliana lines exhibiting enhanced somatic homologous recombination yielded a mutant affected in expression of a gene encoding a caltractin-like protein (centrin). The hyperrecombinogenic phenotype could be reproduced using RNA interference (RNAi) technology. Both the original mutant and the RNAi plants exhibited a moderate UV-C sensitivity as well as a reduced efficiency of in vitro repair of UV-damaged DNA. Transcription profiling of the mutant showed that expression of components of the nucleotide excision repair (NER) pathway and of factors involved in other DNA repair processes were significantly changed. Our data suggest an indirect involvement of centrin in recombinational DNA repair via the modulation of the NER pathway. These findings thus point to a novel interconnection between an early step of NER and homologous recombination, which may play a critical role in plant DNA repair.
Copyright 2004 American Society of Plant Biologists
Figures
Figure 1.
Schematic Representation of the Intermolecular Recombination Substrate. T-DNA of the pGRU'S'G'U' plasmid integrated in the IC9 line, carrying two incomplete partially overlapping uidA regions with 1213-bp homologous sequence in direct orientation. Hpt, hygromycin phosphotransferase gene; G, β-glucuronidase gene; LB, left border; P, 35S promoter of Cauliflower mosaic virus; RB, right border; T, 35S Cauliflower mosaic virus terminator.
Figure 2.
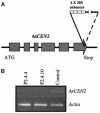
Molecular Analyses of the P2.4 Mutant. (A) Schematic representation of the insertion site of the mutagenic T-DNA upstream of the stop codon of the At4g37010 gene (At_CEN2_). (B) Analysis (RT-PCR) of At4g37010 transcript level in IC9 (control) and in two batches of P2.4 T2 plants. Actin 2 primers were used as control for each PCR reaction.
Figure 3.
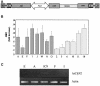
Reproduction of the Hyperrecombinogenic Phenotype by At_CEN2_ Downregulation. (A) Schematic representation of the pOEXhpCEN construct used for downregulation. The fourth exon (156 bp) of the At_CEN2_ gene was cloned into the pOEXhp vector in sense and antisense orientations. LB, left border; mas, mannopine synthase promoter; RB, right border; sul, sulfonamide resistance gene; t, nos terminator. (B) Enhancements of somatic HRF in two independent lines harboring recombination substrates (IC9, closed bars; IC6, dots). Changes of HRF were calculated relative to the corresponding control (IC9 or IC6 plants). One hundred plants were used per replicate, and experiments were duplicated. (C) Expression analysis (RT-PCR) of At_CEN2_ transcript level in IC9 (control) and in four independent RNAi T2 plants. Actin 2 primers were used as control for each PCR reaction.
Figure 4.
Sequence Alignment of At_CEN2_ Gene Product with Six Other Centrins. Comparison of the amino acid sequence of AtCEN2 from Arabidopsis to the three centrin isoforms from human (Hs), to the two isoforms from tobacco (Nt), and to one AtCEN1 from Arabidopsis (At). Identical amino acids are in red, similar amino acids are in orange, and amino acids sharing 30% conservation are in blue. The ATP/GTP binding domain is highlighted in a red rectangle, and the four EF-hand calcium binding sites are denoted by black rectangles.
Figure 5.
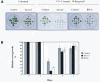
UV-C Sensitivity Assays. (A) One-week-old mutant (At_cen2_), RNAi (pOEXhpCENA), and control plants (IC9) were exposed to a sublethal UV-C dose (30 Kergs/cm2). The phenotype was evaluated 1 week after exposure. (B) Root-growth assay. Three-day-old mutant (At_cen2_), RNAi (pOEXhpCENA), and control plants (IC9) were exposed to 10 Kergs/cm2 of UV-C. Root growth was measured 2 d before (days −2 and −1) and 3 d after irradiation (days 1, 2, and 3). Root growth was calculated relative to the corresponding untreated plants. Ten plants per replicate were used, and experiments were triplicated.
Figure 6.
In Vitro DNA Repair Assay of UV-C–Damaged Plasmid. Cell extracts (25 μg) from mutant (At_cen2_) and control (IC9) plants (A) and from centrin overexpressor and control (wild-type) plants (B) were incubated with UV-C–damaged (UV-C–treated pGEX, +UV-C) and control (untreated pBKS, -UV-C) plasmids in the presence of DIG dUTP. Incorporation was evaluated during a time course.
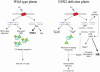
Figure 7.
A Model Explaining the Hyperrecombinogenic Phenotype of At_CEN2_-Deficient Plants. In response to UV irradiation, cyclobutyl pyrimidine dimers, (6-4)-photoproducts (blue trapezium) are formed. In addition, DNA DSBs are introduced. Different repair processes are activated. In At_CEN2_ wild-type plants, DNA photolyase contributes efficiently to the direct repair of UV damage, whereas NER contributes to the dark repair of this damage. For the recognition step of the UV lesion, a complex consisting of RAD4-RAD23 (blue hexagon-green triangle, respectively) is stabilized by CEN2. DNA DSBs can be formed as the result of an extra nick in a single stranded region because of mechanical stress of a chromosome. These DSBs are mainly repaired by the NHEJ (bold), and recombinational DNA repair (HR) remains a minor pathway (lower case). In the At_cen2_-deficient plants, the lower mRNA steady state level of photolyase (lower case) could contribute to a lower efficiency of direct repair of UV damage. In addition, the lack of CEN2 as well as the reduced amount of RAD4 may reduce the stability/efficiency of the recognition complex involved in the first step of the NER pathway. Therefore, the reduced efficiency of the two major UV repair pathways may lead to the accumulation of DSBs. Repair of these is taken over by HR (bold). Concomitantly, the DSB repair process could be activated by the ATM signaling pathway. MIM may also contribute to remodel chromatin during the HR process.
Similar articles
- An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.
Banerjee S, Roy S. Banerjee S, et al. Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review. - CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions.
Liang L, Flury S, Kalck V, Hohn B, Molinier J. Liang L, et al. Plant Mol Biol. 2006 May;61(1-2):345-56. doi: 10.1007/s11103-006-0016-9. Plant Mol Biol. 2006. PMID: 16786311 - Genome stability in the uvh6 mutant of Arabidopsis thaliana.
Bilichak A, Yao Y, Titov V, Golubov A, Kovalchuk I. Bilichak A, et al. Plant Cell Rep. 2014 Jun;33(6):979-91. doi: 10.1007/s00299-014-1580-0. Epub 2014 Feb 20. Plant Cell Rep. 2014. PMID: 24553752 - Arabidopsis homologue of human transcription factor IIH/nucleotide excision repair factor p44 can function in transcription and DNA repair and interacts with AtXPD.
Vonarx EJ, Tabone EK, Osmond MJ, Anderson HJ, Kunz BA. Vonarx EJ, et al. Plant J. 2006 May;46(3):512-21. doi: 10.1111/j.1365-313X.2006.02705.x. Plant J. 2006. PMID: 16623910 - Role of the AtRad1p endonuclease in homologous recombination in plants.
Dubest S, Gallego ME, White CI. Dubest S, et al. EMBO Rep. 2002 Nov;3(11):1049-54. doi: 10.1093/embo-reports/kvf211. Epub 2002 Oct 22. EMBO Rep. 2002. PMID: 12393748 Free PMC article.
Cited by
- Arabidopsis RAD16 Homologues Are Involved in UV Tolerance and Growth.
Alrayes L, Stout J, Schroeder D. Alrayes L, et al. Genes (Basel). 2023 Jul 28;14(8):1552. doi: 10.3390/genes14081552. Genes (Basel). 2023. PMID: 37628604 Free PMC article. - Time-Series Transcriptomic Analysis of Contrasting Rice Materials under Heat Stress Reveals a Faster Response in the Tolerant Cultivar.
Cai H, Wang H, Zhou L, Li B, Zhang S, He Y, Guo Y, You A, Jiao C, Xu Y. Cai H, et al. Int J Mol Sci. 2023 May 28;24(11):9408. doi: 10.3390/ijms24119408. Int J Mol Sci. 2023. PMID: 37298358 Free PMC article. - The nucleotide metabolome of germinating Arabidopsis thaliana seeds reveals a central role for thymidine phosphorylation in chloroplast development.
Niehaus M, Straube H, Specht A, Baccolini C, Witte CP, Herde M. Niehaus M, et al. Plant Cell. 2022 Sep 27;34(10):3790-3813. doi: 10.1093/plcell/koac207. Plant Cell. 2022. PMID: 35861422 Free PMC article. - Structural Basis for the Functional Diversity of Centrins: A Focus on Calcium Sensing Properties and Target Recognition.
Pedretti M, Bombardi L, Conter C, Favretto F, Dominici P, Astegno A. Pedretti M, et al. Int J Mol Sci. 2021 Nov 10;22(22):12173. doi: 10.3390/ijms222212173. Int J Mol Sci. 2021. PMID: 34830049 Free PMC article. Review. - An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.
Banerjee S, Roy S. Banerjee S, et al. Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
References
- Araki, M., Masutani, C., Takemura, M., Uchida, A., Sugasawa, K., Kondoh, J., Ohkuma, Y., and Hanaoka, F. (2001). Centrosome protein centrin 2/caltractin 1 is part of the Xeroderma Pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 276, 18665–18672. - PubMed
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. - PubMed
- Britt, A.B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant Sci. 4, 20–24. - PubMed
- Cole, R.S., and Sinden, R.R. (1975). Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 5, 487–495. - PubMed
- Cordeiro, M.C.R., Piqueras, R., de Oliveira, D.E., and Catresana, C. (1998). Characterization of early induced genes in Arabidopsis thaliana responding to bacterial inoculation: Identification of centrin and of a novel protein with two regions related to kinase domains. FEBS Lett. 434, 387–393. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases