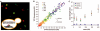Control of stochasticity in eukaryotic gene expression - PubMed (original) (raw)
Control of stochasticity in eukaryotic gene expression
Jonathan M Raser et al. Science. 2004.
Abstract
Noise, or random fluctuations, in gene expression may produce variability in cellular behavior. To measure the noise intrinsic to eukaryotic gene expression, we quantified the differences in expression of two alleles in a diploid cell. We found that such noise is gene-specific and not dependent on the regulatory pathway or absolute rate of expression. We propose a model in which the balance between promoter activation and transcription influences the variability in messenger RNA levels. To confirm the predictions of our model, we identified both cis- and trans-acting mutations that alter the noise of gene expression. These mutations suggest that noise is an evolvable trait that can be optimized to balance fidelity and diversity in eukaryotic gene expression.
Figures
Fig. 1
Separation of intrinsic and extrinsic noise for the PHO5 promoter. (A) A false-color overlay of YFP (red) and CFP (green) fluorescence micrographs from a diploid yeast strain that expresses YFP and CFP from identical promoters at homologous loci, as diagrammed in the inset. (B) Scatter plots showing CFP and YFP values for each cell (solid circles) during a time course of PHO5 induction by phosphate starvation. Populations from different time points (in minutes) are indicated with different colors. Extrinsic noise is manifested as scatter along the diagonal and intrinsic noise as scatter perpendicular to the diagonal. AU, arbitrary units of fluorescence. (C) Total, extrinsic, and intrinsic noise strength as functions of population mean for (B). The solid line represents expectations for a single stochastic process, and error bars represent bootstrap values (6).
Fig. 2
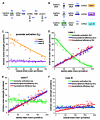
(A) Intrinsic noise strength as a function of rate of expression for GAL1 (left) and a scatter plot of _GAL1_-expressing cells at maximal induction (right). To produce different rates of expression, cells were induced with different galactose concentrations. (B) Intrinsic noise strength as a function of rate of expression for PHO84 and a scatter plot of _PHO84_-expressing cells at maximal induction. Cells were induced with different phosphate concentrations. (C) Intrinsic noise strength as a function of rate of expression for PHO5 and a scatter plot of _PHO5_-expressing cells at the maximal level of induction by phosphate starvation. Cells were induced with various levels of chemical inhibition of an upstream kinase (left) or with various organic phosphate concentrations (inset). The dashed line indicates the intrinsic error of measurement, and error bars represent standard deviations. Scatter plots contain cells from multiple time points.
Fig. 3
General stochastic model of gene activation and expression. (A) Schematic of reactions with stochastic rate constants of production (k) and degradation (γ). Ø indicates the null product of degradation. a, active DNA; m, mRNA; p, protein. (B) Three cases, where the relative size of the arrows indicates the relative magnitude of the constants within each case. (C) The effect on intrinsic noise strength of changing the promoter activation rate to change the steady-state mean of expression for case I (orange □), case II (violet □), and case III (cyan □). (D to F) The effect on intrinsic noise strength of changing promoter activation (green □), transcriptional efficiency (red ○), and translational efficiency (purple △) to change the steady-state mean of expression for (D) case I, (E) case II, and (F) case III. For (C) to (F), the solid lines display the predicted values, and the open symbols are averages from stochastic simulations (6).
Fig. 4
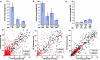
Mutational analysis of the PHO5 promoter. (A) Intrinsic noise strength and rate of expression of wild-type, UASm1, and UASm2 PHO5 promoter variants at maximal induction (top) and a scatter plot for the wild type (○) and UASm1 (red ◆) (bottom). The intrinsic noise of the UASm1 promoter is 77% at a mean of 26 AU, compared to 43% intrinsic noise at a mean of 29 AU for the wild-type promoter. (B) Intrinsic noise strength and rate of expression of the PHO5 promoter at maximal induction in the wild-type background or in strains that lack SNF6, ARP8, or GCN5 (top) and a scatter plot for the wild type (○) and _snf6_Δ (red ◆) (bottom). The intrinsic noise of the _snf6_Δ strain is 73% at a mean of 35 AU. (C) Intrinsic noise strength and rate of expression of wild-type and TATA mutant PHO5 promoters at maximal induction (top) and a scatter plot for the wild type (○) and TATA-C2 (red ◆) (bottom). The intrinsic noise of the TATA-C2 promoter is 15% at a mean of 25 AU. Error bars represent standard deviations. Scatter plots contain cells from multiple time points.
Similar articles
- Noise in eukaryotic gene expression.
Blake WJ, KAErn M, Cantor CR, Collins JJ. Blake WJ, et al. Nature. 2003 Apr 10;422(6932):633-7. doi: 10.1038/nature01546. Nature. 2003. PMID: 12687005 - Stochastic gene expression in a single cell.
Elowitz MB, Levine AJ, Siggia ED, Swain PS. Elowitz MB, et al. Science. 2002 Aug 16;297(5584):1183-6. doi: 10.1126/science.1070919. Science. 2002. PMID: 12183631 - Contributions of low molecule number and chromosomal positioning to stochastic gene expression.
Becskei A, Kaufmann BB, van Oudenaarden A. Becskei A, et al. Nat Genet. 2005 Sep;37(9):937-44. doi: 10.1038/ng1616. Epub 2005 Aug 7. Nat Genet. 2005. PMID: 16086016 - The role of chromatin structure in regulating stress-induced transcription in Saccharomyces cerevisiae.
Uffenbeck SR, Krebs JE. Uffenbeck SR, et al. Biochem Cell Biol. 2006 Aug;84(4):477-89. doi: 10.1139/o06-079. Biochem Cell Biol. 2006. PMID: 16936821 Review. - Noise in gene expression: origins, consequences, and control.
Raser JM, O'Shea EK. Raser JM, et al. Science. 2005 Sep 23;309(5743):2010-3. doi: 10.1126/science.1105891. Science. 2005. PMID: 16179466 Free PMC article. Review.
Cited by
- A DNA base-specific sequence interposed between CRX and NRL contributes to RHODOPSIN expression.
Maritato R, Medugno A, D'Andretta E, De Riso G, Lupo M, Botta S, Marrocco E, Renda M, Sofia M, Mussolino C, Bacci ML, Surace EM. Maritato R, et al. Sci Rep. 2024 Nov 1;14(1):26313. doi: 10.1038/s41598-024-76664-8. Sci Rep. 2024. PMID: 39487168 Free PMC article. - Altruistic feeding and cell-cell signaling during bacterial differentiation actively enhance phenotypic heterogeneity.
Updegrove TB, Delerue T, Anantharaman V, Cho H, Chan C, Nipper T, Choo-Wosoba H, Jenkins LM, Zhang L, Su Y, Shroff H, Chen J, Bewley CA, Aravind L, Ramamurthi KS. Updegrove TB, et al. Sci Adv. 2024 Oct 18;10(42):eadq0791. doi: 10.1126/sciadv.adq0791. Epub 2024 Oct 18. Sci Adv. 2024. PMID: 39423260 Free PMC article. - MONITTR allows real-time imaging of transcription and endogenous proteins in C. elegans.
Liu X, Chang Z, Sun P, Cao B, Wang Y, Fang J, Pei Y, Chen B, Zou W. Liu X, et al. J Cell Biol. 2025 Jan 6;224(1):e202403198. doi: 10.1083/jcb.202403198. Epub 2024 Oct 14. J Cell Biol. 2025. PMID: 39400293 - Monosome Stalls the Translation Process Mediated by IGF2BP in Arcuate Nucleus for Puberty Onset Delay.
Shen Y, Zhang L, Yang T, Li X, Liu C, Li H, Hu Y, Shen H, Li H, Orlov YL, Zhou S, Shen Y. Shen Y, et al. Mol Neurobiol. 2024 Sep 5. doi: 10.1007/s12035-024-04450-8. Online ahead of print. Mol Neurobiol. 2024. PMID: 39235646 - Transcriptional noise, gene activation, and roles of SAGA and Mediator Tail measured using nucleotide recoding single-cell RNA-seq.
Schofield JA, Hahn S. Schofield JA, et al. Cell Rep. 2024 Aug 27;43(8):114593. doi: 10.1016/j.celrep.2024.114593. Epub 2024 Aug 4. Cell Rep. 2024. PMID: 39102335 Free PMC article.
References
- McAdams HH, Arkin A. Trends Genet. 1999;15:65. - PubMed
- Schroedinger E. What Is Life? Cambridge Univ. Press; Cambridge: 1944.
- Rao CV, Wolf DM, Arkin AP. Nature. 2002;420:231. - PubMed
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials