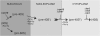The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast - PubMed (original) (raw)
The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast
Isabelle Léger-Silvestre et al. EMBO J. 2004.
Abstract
We have conducted a genetic screen in order to identify ribosomal proteins of Saccharomyces cerevisiae involved in nuclear export of the small subunit precursors. This has led us to distinguish Rps15p as a protein dispensable for maturation of the pre-40S particles, but whose assembly into the pre-ribosomes is a prerequisite to their nuclear exit. Upon depletion of Rps15p, 20S pre-rRNA is released from the nucleolus and retained in the nucleus, without alteration of the pre-rRNA early cleavages. In contrast, Rps18p, which contacts Rps15p in the small subunit, is required upstream for pre-rRNA processing at site A2. Most pre-40S specific factors are correctly associated with the intermediate particles accumulating in the nucleus upon Rps15p depletion, except the late-binding proteins Tsr1p and Rio2p. Here we show that these two proteins are dispensable for nuclear exit; instead, they participate in 20S pre-rRNA processing in the cytoplasm. We conclude that, during the final maturation steps in the nucleus, incorporation of the ribosomal protein Rps15p is specifically required to render the pre-40S particles competent for translocation to the cytoplasm.
Figures
Figure 1
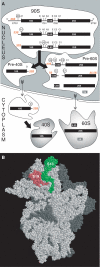
Ribosome biogenesis. (A) Pre-rRNA processing and nuclear export in S. cerevisiae. (B) Structure of the T. thermophilus 30S subunit. The ribosomal proteins S19 (red) and S13 (green), homologous to the eukaryotic proteins Rps15p and Rps18p, respectively, form a dimer on the verge of the interface domain. The other ribosomal proteins are shown in grey and the 16S rRNA in white. Figure derived from PDB file 1J5E (Wimberly et al, 2000) using RasMol.
Figure 2
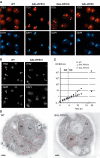
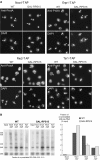
Rps15p-depleted cells accumulate precursors to the 18S rRNA in the nucleus. (A) Pre-18S rRNA FISH with a probe complementary to the D–A2 segment of the ITS1 in wild-type, GAL-RPS15, GAL-RPS18, and GAL-RPS0 strains grown for 4 h in glucose-containing medium. Arrowheads indicate the nucleoplasm as visualized by DNA staining with DAPI. (B) Pre-5.8/25S rRNA detected with a probe to the E-C2 domain in the ITS2. (C) Growth of wild-type, GAL-RPS15, and GAL RPS18 cells. At _t_=4.5 h (dotted line), YP-galactose medium was replaced by YP-glucose. (D) Detection by electron microscopy of pre-18S RNAs with a riboprobe complementary to the D–A2 fragment of the ITS1. Cells were grown as in (A). Gold particles are highlighted in red. No: nucleolus; Np: nucleoplasm; Cy: cytoplasm. Bar=200 nm.
Figure 3
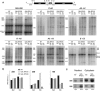
Northern blot analysis of pre-rRNA processing in Gal-RPS15 and GAL-RPS18 cells. (A) Northern blot of total RNA extracts from wild-type, GAL-RPS15, and GAL-RPS18 cells grown in YP-galactose medium (Gal), or transferred for 4 or 8 h to YP-glucose (Glc). (B) Levels of 25S, 20S, and 18S RNAs in the cells grown in YP-galactose medium (Gal) or shifted to YP-glucose medium for 4 h (Glc); quantitative analysis was performed on the Northern blots probed with the 25S, 18S, and D–A2 probes (a.u.: arbitrary units). (C) Detection of the 20S and 25S RNAs extracted from the nuclear and cytoplasmic fractions of wild-type and GAL-RPS15 cells grown for 4 h in YP-glucose medium. The distribution of 20S pre-rRNA in the two fractions was evaluated by phosphorimager quantification.
Figure 4
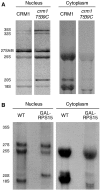
Analysis of newly synthesized pre-ribosome nuclear export by metabolic labelling and cell fractionation. Newly synthesized RNAs in spheroblasts were labelled for 15 min with [3H]uracil prior to nuclear-cytoplasmic fractionation. RNAs were then isolated and analysed by gel electrophoresis and fluorography. Equivalent amounts of radioisotope were loaded in each lane. (A) Nuclear export of pre-ribosomes in _crm1_Δ cells expressing either the wild-type CRM1 gene (MNY7) or the leptomycin B (LMB)-sensitive crm1T539C mutant allele (MNY8) from a plasmid. LMB was added to the medium 5 min before the addition of [3H]uracil. Note the nuclear retention of the 25S and 20S (pre-) rRNA in the LMB-sensitive cells. (B) Upon Rps15p depletion in GAL-RPS15 cells (4 h in the presence of glucose), 20S pre-rRNA, but not 25S rRNA, is blocked in the nucleus.
Figure 5
Behaviour of pre-ribosomal factors in Rps15p-depleted cells. (A) Immunolocalization of TAP-tagged Noc4p, Enp1p, Tsr1p, and Rio2p in wild-type and GAL-RPS15 strains cultured for 4 h in the presence of glucose. Immunofluorescence with anti-protein A antibodies. Arrowheads indicate the nucleoplasm as visualized by DNA staining with DAPI. (B) Detection of pre-18S rRNAs co-purifying with TAP-tagged Noc4p, Enp1p, Rio2p, and Tsr1p in Rps15p-depleted cells. GAL-RPS15 cells expressing one of the TAP-tagged proteins were grown for 4 h in YP-glucose medium and total cell extracts were submitted to immunoprecipitation with IgG-Sepharose. Co-precipitating RNAs were revealed after Northern blot with a probe hybridizing between points D and A2 in the ITS1. All samples, except Tsr1-TAP, were processed in parallel, starting with equivalent amounts of cell extract; one representative input lane (0.05% of input) is displayed. Semiquantification of the fraction of precipitated RNA was performed by phosphorimager analysis.
Figure 6
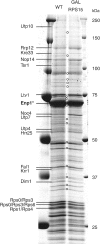
Composition of the pre-40S complexes in wild-type and Rps15p-depleted cells. Wild-type and GAL-RPS15 cells expressing Enp1-TAP were grown in the presence of galactose and then shifted to medium containing glucose for 3 h. After TAP purification, the Enp1p-associated proteins were separated by SDS–PAGE and identified by mass spectrometry. Rio2p could not be detected in these preparations, and may be masked by a degradation product of Enp1p-TAP.
Figure 7
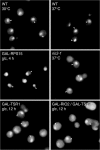
Rio2p and Tsr1p are required for 20S pre-rRNA cleavage in the cytoplasm. Cells bearing the thermosensitive allele rio2-1 were grown in YP-glucose medium at 25°C and shifted to 37°C for 3 h. GAL-TSR1 cells were grown in glucose-containing medium for 12 h. ITS1 detection by FISH shows a clear increase of the cytoplasmic signal under nonpermissive conditions, compared to wild-type cells. This phenotype is also observed when Rio2p and Tsr1p are simultaneously affected in a GAL-RIO2/GAL-TSR1 strain. GAL-RPS15 cells grown under nonpermissive condition are presented here for comparison. Arrowheads indicate position of the nucleoplasm as detected in parallel by DAPI staining.
Figure 8
Involvement of Rps15p, Rps18p, and the late pre-ribosomal factors Tsr1p and Rio2p in the small subunit biogenesis pathway. Once released from the nucleolus, the pre-40S particles become competent for nuclear export (pre-40S′), a process that requires at least Rps15p and Crm1p. After translocation to the cytoplasm, association of the Rio proteins precedes the conversion to mature 40S subunits. Tsr1p may be incorporated earlier, in the nucleus (see Discussion).
Similar articles
- Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae.
Vanrobays E, Gelugne JP, Gleizes PE, Caizergues-Ferrer M. Vanrobays E, et al. Mol Cell Biol. 2003 Mar;23(6):2083-95. doi: 10.1128/MCB.23.6.2083-2095.2003. Mol Cell Biol. 2003. PMID: 12612080 Free PMC article. - Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals.
Carron C, O'Donohue MF, Choesmel V, Faubladier M, Gleizes PE. Carron C, et al. Nucleic Acids Res. 2011 Jan;39(1):280-91. doi: 10.1093/nar/gkq734. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805244 Free PMC article. - Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast.
Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Gelperin D, et al. RNA. 2001 Sep;7(9):1268-83. doi: 10.1017/s1355838201013073. RNA. 2001. PMID: 11565749 Free PMC article. - Nuclear export and cytoplasmic maturation of ribosomal subunits.
Zemp I, Kutay U. Zemp I, et al. FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review. - Maturation of pre-40S particles in yeast and humans.
Cerezo E, Plisson-Chastang C, Henras AK, Lebaron S, Gleizes PE, O'Donohue MF, Romeo Y, Henry Y. Cerezo E, et al. Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1516. doi: 10.1002/wrna.1516. Epub 2018 Nov 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30406965 Review.
Cited by
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation.
Murgiano L, Shirokova V, Welle MM, Jagannathan V, Plattet P, Oevermann A, Pienkowska-Schelling A, Gallo D, Gentile A, Mikkola M, Drögemüller C. Murgiano L, et al. PLoS Genet. 2015 Jul 23;11(7):e1005427. doi: 10.1371/journal.pgen.1005427. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26203908 Free PMC article. - The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome.
Kühn H, Hierlmeier T, Merl J, Jakob S, Aguissa-Touré AH, Milkereit P, Tschochner H. Kühn H, et al. PLoS One. 2009 Dec 18;4(12):e8370. doi: 10.1371/journal.pone.0008370. PLoS One. 2009. PMID: 20019888 Free PMC article. - Good Vibrations: Structural Remodeling of Maturing Yeast Pre-40S Ribosomal Particles Followed by Cryo-Electron Microscopy.
Shayan R, Rinaldi D, Larburu N, Plassart L, Balor S, Bouyssié D, Lebaron S, Marcoux J, Gleizes PE, Plisson-Chastang C. Shayan R, et al. Molecules. 2020 Mar 3;25(5):1125. doi: 10.3390/molecules25051125. Molecules. 2020. PMID: 32138239 Free PMC article. - Multiomic analysis of Schistosoma mansoni reveals unique expression profiles in cercarial heads and tails.
Hagerty JR, Kim HC, Jolly ER. Hagerty JR, et al. Commun Biol. 2021 Jul 12;4(1):860. doi: 10.1038/s42003-021-02366-w. Commun Biol. 2021. PMID: 34253841 Free PMC article. - Ribosomal protein uS19 mutants reveal its role in coordinating ribosome structure and function.
Bowen AM, Musalgaonkar S, Moomau CA, Gulay SP, Mirvis M, Dinman JD. Bowen AM, et al. Translation (Austin). 2015 Nov 18;3(2):e1117703. doi: 10.1080/21690731.2015.1117703. eCollection 2015 Jul-Dec. Translation (Austin). 2015. PMID: 26824029 Free PMC article.
References
- Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V (2002) Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316: 725–768 - PubMed
- Fatica A, Tollervey D (2002) Making ribosomes. Curr Opin Cell Biol 14: 313–318 - PubMed
- Ford CL, Randal-Whitis L, Ellis SR (1999) Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res 59: 704–710 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases