A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein - PubMed (original) (raw)
A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein
Juan Carlos Arévalo et al. EMBO J. 2004.
Abstract
A major question in cell biology is how molecular specificity is achieved by different growth factor receptors that activate apparently identical signaling events. For the neurotrophin family, a distinguishing feature is the ability to maintain a prolonged duration of signal transduction. However, the mechanisms by which neurotrophin receptors assemble such a sustained signaling complex are not understood. Here we report that an unusual ankyrin-rich transmembrane protein (ARMS+kidins220) is closely associated with Trk receptor tyrosine kinases, and not the EGF receptor. This association requires interactions between transmembrane domains of Trk and ARMS. ARMS is rapidly tyrosine phosphorylated after binding of neurotrophins to Trk receptors and provides a docking site for the CrkL-C3G complex, resulting in Rap1-dependent sustained ERK activation. Accordingly, disruption of Trk-ARMS or the ARMS-CrkL interaction with dominant-negative ARMS mutants, or treatment with small interference RNA against ARMS substantially reduce neurotrophin-elicited signaling to ERK, but without any effect upon Ras or Akt activation. These findings suggest that ARMS acts as a major and neuronal-specific platform for prolonged MAP kinase signaling by neurotrophins.
Figures
Figure 1
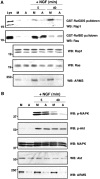
ARMS is rapidly tyrosine phosphorylated upon neurotrophin treatment. (A) Kinetics of TrkA signaling. PC12 cells were treated with NGF (100 ng/ml) for the indicated time (minutes). Immunoprecipitation was performed for Trk, ARMS, PLC-γ and Shc, followed by Western blot with 4G10, an anti-phosphotyrosine antibody, whereas 50 μg of whole lysates was used to detect the phosphorylation status of MAPK and Akt using phosphospecific antibodies (left panel). The same blot was re-probed with Trk, ARMS, PLC-γ, Shc, MAPK and Akt antibodies to verify the equality of protein loading (right panel). (B) Kinetics of TrkB signaling. Cortical neurons (8–10 DIV) treated with BDNF (50 ng/ml) were added for the indicated time (s). Immunoprecipitation was carried out as described in (A). The left blot was probed with 4G10 and the total amount of proteins is shown in the right panel.
Figure 2
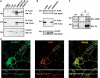
ARMS and Trk receptors interact. (A) ARMS interacts with Trk receptors. Lysates from HEK293 cells transfected with FLAG-ARMS and/or TrkA, B, or C receptor were immunoprecipitated with FLAG antibodies, followed by Western blotting by anti-Trk antisera. (B) Immunoprecipitated ARMS does not bind EGF receptors. Experiments were carried out in the same way as described in panel A, but EGF receptor was analyzed. (C) ARMS interacts with TrkB in primary cortical neurons. Cells were cultured for 8–12 DIV and crosslinked with the lipid-soluble compound DSP (Pierce), a thiol-cleavable reagent. Control rabbit IgG was used instead of anti-Trk to demonstrate the specificity of the Trk–ARMS interaction. (D) Co-localization of TrkB and ARMS in primary cortical neurons. Staining was carried out with affinity-purified rabbit polyclonal antibody 892 against ARMS (green; Kong et al, 2001) and the mouse monoclonal antibody B3 (Santa Cruz) against TrkB (red). Co-localization of both proteins is shown in yellow. Images were collected on a confocal microscope. An area within the cortical neuron was enlarged to highlight TrkB and ARMS colocalization. Scale bar=10 μm.
Figure 3
Transmembrane domains are responsible for the interaction between Trk receptors and ARMS. (A) Transmembrane region of TrkA is necessary for the interaction with ARMS. Anti-FLAG immunoprecipitation was performed with extracts from HEK293 cells transfected with FLAG-ARMS and wild-type TrkA or a chimera of TrkA/EGFR that contains the transmembrane region of the EGF receptor. The presence of Trk was verified by Western blotting with an anti-Trk antisera. (B) Summary of Trk/ARMS interaction using different Trk mutants. The regions deleted were the juxtamembrane (ΔJM), extracellular (ΔECD) or intracellular (ΔICD) domain (Arévalo et al, 2000; Yano et al, 2000, 2001). The chimera TrkA-TM EGF receptor was generated by swapping the transmembrane region of TrkA for the corresponding domain from the EGF receptor. See Supplementary data for ARMS binding to Trk receptor mutants (Supplementary Figure S2). (C) Lack of transmembrane regions in ARMS abolished the interaction with TrkA. Anti-FLAG immunoprecipitation experiments were performed with extracts from HEK293 cells transfected with wild-type TrkA and FLAG-tagged full-length ARMS or truncated ARMS, followed by Western blotting with an anti-Trk antisera. Arrows indicate the migration of different truncated ARMS proteins. (D) Summary of TrkA/ARMS interaction using different ARMS mutants. Similar experiments as in Figure 4C were carried out to identify the ARMS regions involved in the TrkA interaction. TM=transmembrane domain; Pro=polyproline-rich region; SAM=sterile α-motif; PDZ=binding motif (SIL). See Supplementary data for analysis of TrkA and ARMS mutants (Supplementary Figure S3). (E) A truncated form of ARMS impairs the interaction of TrkA and full-length ARMS. Extracts from HEK293 cells transfected with full-length FLAG-ARMS, TrkA and increasing concentrations of FLAG-ARMS4.2 were immunoprecipitated with anti-TrkA antibodies (Clary et al, 1994) and Western blot were performed with anti-FLAG antibody to detect ARMS and ARMS4.2 expression. Note that increasing levels of ARMS4.2 diminish TrkA interaction with full-length ARMS.
Figure 4
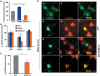
ARMS4.2 interferes with sustained MAPK phosphorylation. (A) Neurite outgrowth of PC12 cells transfected with GFP, FLAG-ARMS or FLAG-ARMS4.2. Cells were stimulated with NGF for 24 h after transfection and neurite outgrowth was quantified 3 days later by assessing the percentage of fluorescence-positive cells bearing neurites at least twice the length of the cell body. At least 50 cells were counted for each experiment and condition. Results were normalized to control GFP-transfected cells (set as 100%). Values are calculated from at least four independent experiments. Mean and standard deviation (s.d.) are shown. Where applicable, statistical analyses were carried out by Student's _t_-test (*P<0.02; **P<0.001). (B) Activation of p-MAPK in PC12 cells transfected with GFP, FLAG-ARMS or FLAG-ARMS4.2. Respective cultures were stimulated with NGF for 5 or 40 min or EGF for 5 min. Quantification was carried out as described in panel A (**P<0.001). (C) Immunofluorescence analysis of primary cortical neurons transfected with ARMS4.2 constructs. Cells were transfected with GFP or GFP/FLAG-ARMS4.2 and stimulated 2 days later with BDNF for 40 min. Panel b shows the absence of p-MAPK staining in untreated cells. Panels d–l show transfected neurons with GFP and GFP/FLAG-ARMS4.2. Panels d–f and g–l show GFP and GFP/FLAG-ARMS4.2-transfected neurons that are positive (yellow, panel f) and negative (green, panel i) for p-MAPK, respectively. Panels j–l show GFP/FLAG-ARMS4.2-transfected neurons stained for p-Akt (yellow, panel l). Scale bar=5 μm. (D) Activation of p-MAPK in cortical neurons transfected with GFP and GFP/ARMS4.2 stimulated with BDNF for 40 min (**P<0.001).
Figure 5
TrkA, ARMS and CrkL association is enhanced by NGF. (A) Deletion of the Shc-binding site abolished NGF-induced FRS-2 interaction, but did not affect complex formation between TrkA and CrkL/c-Crk II. PC12nnr5 cells stably expressing wild-type TrkA or a mutant Trk (ΔShc) carrying a deletion (ENPQY490F) that does not bind Shc or FRS2 were treated with NGF for 10 min. Lysates were immunoprecipitated either with an anti-Crk antibody, followed by blotting with an anti-Trk antiserum (RTA), or with anti-FRS2 antiserum and blotting with antiphosphotyrosine (p-Tyr). Reprobing the blots with anti-CrkL and anti-FRS2 verified sample loading. (B) TrkA, ARMS and CrkL association in PC12 cells is enhanced by NGF treatment. PC12-615 cells were treated for the indicated times with 100 ng/ml NGF. Detergent lysates were immunoprecipitated with anti-ARMS polyclonal antibody. Western blotting was performed with Trk (C-14) and CrkL polyclonal antibodies. (C) CrkL increases the binding of C3G and Trk upon NGF treatment. PC12-615 cells were stimulated for the indicated time with NGF and extracts were immunoprecipitated with pan-Crk antibodies. Western blotting analysis was performed with C3G, Abl, CrkL and TrkA to detect these proteins. Note that the increasing levels of C3G and activated TrkA were pulled down together with CrkL in response to NGF treatment. (D) Association of Trk, ARMS and CrkL. HEK293T cells were transfected with the indicated plasmids. Anti-FLAG immunoprecipitates were blotted with the indicated antibodies. The association of TrkA with CrkL was enhanced by ARMS expression (lane 1 versus lane 4).
Figure 6
Ligand dependency of ARMS, CrkL and C3G interactions. (A) The polyproline-rich region of ARMS contains consensus sequences (PXXP) for binding to SH3 domains. Sequences within the polyproline-rich region of ARMS (P1057–L1151) present three PXXP motifs (bold) that are consensus for binding of SH3 domain-containing molecules (Feller, 2001) and are predicted by website sequence analysis:
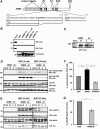
. (B) In vitro interaction between ARMS and CrkL, but not c-CrkII. GST–ARMS recombinant proteins were incubated with HEK293 cell extracts and subjected to Western blotting analysis with anti-c-Crk II or anti-CrkL antisera. A Coomassie-stained gel of the input GST fusion proteins is shown (bottom panel). (C) PC12 cells were stably transfected with FLAG-ARMS and FLAG-ARMSΔP. Individual clones were analyzed for the ectopic expression of ARMS or ARMSΔP proteins using anti-FLAG antibodies. Reprobing the blot with anti-ARMS antibody revealed that the overexpression levels for Flag-ARMS and Flag-ARMSΔP are approximately three-fold. (D) Expression of ARMSΔP abolishes Rap1 but not Ras activation elicited by NGF. PC12 clones stably expressing FLAG-ARMS or FLAG-ARMSΔP were serum starved for 16 h, lysates were prepared and subjected to incubation with 10 μg of GST-RalGDS RBD or the Raf-1 Ras-binding domain protein (GST-Raf RBD) in pull-down assays (Hermann et al, 1996) to detect active Rap1 and Ras, respectively. Immunoblots using anti-Rap1 and anti-Ras were carried out as indicated. (E) Expression of FLAG-ARMSΔP in PC12 cells impairs sustained MAP kinase activation elicited by NGF. Cell extracts from PC12 cells or PC12 clones described above (C), nontreated or treated with NGF for 5 or 40 min, were obtained and Western blots were performed with p-MAPK and p-Akt antibodies. A reduction in MAPK activation in PC12 clones expressing FLAG-ARMSΔP was seen at 40 min of NGF treatment, but no differences were observed in the activation of Akt. (F) Neurite outgrowth of PC12 cells transiently transfected with GFP, FLAG-ARMS or FLAG-ARMSΔP. At 24 h after transfection with the indicated plasmids, cells were treated with NGF for 2 additional days. PC12 transfectants were scored as positive as described in Figure 4A (*P<0.02; **P<0.001). (G) Cortical neurons transfected with GFP/FLAG-ARMSΔP showed a reduced response to BDNF. Cells were transfected with GFP or GFP/FLAG-ARMSΔP. After 2 days, cultures were stimulated with BDNF for 40 min and staining for p-MAPK was performed. The statistical significance by Student's _t_-test was demonstrated for (F) and (G) (*P<0.02; **P<0.001).
Figure 7
Downregulation of ARMS expression by siRNA impairs Rap1 and prolonged MAPK activation in PC12 cells upon NGF treatment. (A) ARMS siRNA impairs Rap1 but not Ras activation upon NGF treatment of PC12 cells. ARMS siRNA was produced as described in Materials and methods. PC12 cells were either mock transfected or transfected with ARMS siRNA. NGF treatment was applied for 5 or 40 min. Equal amounts of cellular lysates were subjected to Ras or Rap1 pulldown, as described in Figure 6D. Total lysates under each treatment condition were subjected to Western blotting with the indicated antibodies. (B) siRNA downregulates ARMS in PC12 cells and specifically decreases the Rap1-dependent prolonged MAPK activation elicited by NGF. Cellular extracts from control or ARMS siRNA-transfected PC12 cells were Western blotted with p-MAPK or pAkt antibody. The levels of ARMS protein in siRNA-transfected cells were assessed by blotting with anti-ARMS (892). A decrease of p-MAPK signal was detected in the cells with a reduced amount of ARMS at 40 min of NGF treatment, but not at 5 min. M, mock; A, ARMS siRNA.
Similar articles
- Identification of a switch in neurotrophin signaling by selective tyrosine phosphorylation.
Arévalo JC, Pereira DB, Yano H, Teng KK, Chao MV. Arévalo JC, et al. J Biol Chem. 2006 Jan 13;281(2):1001-7. doi: 10.1074/jbc.M504163200. Epub 2005 Nov 11. J Biol Chem. 2006. PMID: 16284401 - Ternary complex with Trk, p75, and an ankyrin-rich membrane spanning protein.
Chang MS, Arevalo JC, Chao MV. Chang MS, et al. J Neurosci Res. 2004 Oct 15;78(2):186-92. doi: 10.1002/jnr.20262. J Neurosci Res. 2004. PMID: 15378608 - An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors.
Kong H, Boulter J, Weber JL, Lai C, Chao MV. Kong H, et al. J Neurosci. 2001 Jan 1;21(1):176-85. doi: 10.1523/JNEUROSCI.21-01-00176.2001. J Neurosci. 2001. PMID: 11150334 Free PMC article. - Distinctive features of Trk neurotrophin receptor transactivation by G protein-coupled receptors.
Lee FS, Rajagopal R, Chao MV. Lee FS, et al. Cytokine Growth Factor Rev. 2002 Feb;13(1):11-7. doi: 10.1016/s1359-6101(01)00024-7. Cytokine Growth Factor Rev. 2002. PMID: 11750876 Review. - Promoting neurotrophic effects by GPCR ligands.
Jeanneteau F, Chao MV. Jeanneteau F, et al. Novartis Found Symp. 2006;276:181-9; discussion 189-92, 233-7, 275-81. Novartis Found Symp. 2006. PMID: 16805430 Review.
Cited by
- Rare variants in the neurotrophin signaling pathway implicated in schizophrenia risk.
Kranz TM, Goetz RR, Walsh-Messinger J, Goetz D, Antonius D, Dolgalev I, Heguy A, Seandel M, Malaspina D, Chao MV. Kranz TM, et al. Schizophr Res. 2015 Oct;168(1-2):421-8. doi: 10.1016/j.schres.2015.07.002. Epub 2015 Jul 26. Schizophr Res. 2015. PMID: 26215504 Free PMC article. - Brain-derived neurotrophic factor regulates cholesterol metabolism for synapse development.
Suzuki S, Kiyosue K, Hazama S, Ogura A, Kashihara M, Hara T, Koshimizu H, Kojima M. Suzuki S, et al. J Neurosci. 2007 Jun 13;27(24):6417-27. doi: 10.1523/JNEUROSCI.0690-07.2007. J Neurosci. 2007. PMID: 17567802 Free PMC article. - The ankyrin repeat-rich membrane spanning (ARMS)/Kidins220 scaffold protein is regulated by activity-dependent calpain proteolysis and modulates synaptic plasticity.
Wu SH, Arévalo JC, Neubrand VE, Zhang H, Arancio O, Chao MV. Wu SH, et al. J Biol Chem. 2010 Dec 24;285(52):40472-8. doi: 10.1074/jbc.M110.171371. Epub 2010 Oct 13. J Biol Chem. 2010. PMID: 20943655 Free PMC article. - PKD1, PKD2, and their substrate Kidins220 regulate neurotensin secretion in the BON human endocrine cell line.
Li J, Chen LA, Townsend CM Jr, Evers BM. Li J, et al. J Biol Chem. 2008 Feb 1;283(5):2614-21. doi: 10.1074/jbc.M707513200. Epub 2007 Nov 29. J Biol Chem. 2008. PMID: 18048355 Free PMC article. - Reciprocal regulation of presynaptic and postsynaptic proteins in bipolar spiral ganglion neurons by neurotrophins.
Flores-Otero J, Xue HZ, Davis RL. Flores-Otero J, et al. J Neurosci. 2007 Dec 19;27(51):14023-34. doi: 10.1523/JNEUROSCI.3219-07.2007. J Neurosci. 2007. PMID: 18094241 Free PMC article.
References
- Atwal J, Massie B, Miller F, Kaplan D (2000) The TrkB-Shc site signals neuornal survival and local axon growth via MEK and PI3-kinase. Neuron 27: 265–277 - PubMed
- Barbacid M (1994) The trk family of neurotrophin receptors. J Neurobiol 25: 1386–1403 - PubMed
- Bibel M, Barde Y (2000) Neurotrophins: key regulators of cell fate and cell shape in the vertebrate nervous system. Genes Dev 14: 2919–2937 - PubMed
- Blum R, Kafitz K, Konnerth A (2002) Neurotrophin-evoked depolarization requires the sodium channel NaV1.9. Nature 419: 687–693 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HD023315/HD/NICHD NIH HHS/United States
- P01 HD023315/HD/NICHD NIH HHS/United States
- NS21072/NS/NINDS NIH HHS/United States
- R01 NS021072/NS/NINDS NIH HHS/United States
- R56 NS021072/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous