Reconstitution of a minimal mtDNA replisome in vitro - PubMed (original) (raw)
Reconstitution of a minimal mtDNA replisome in vitro
Jenny A Korhonen et al. EMBO J. 2004.
Abstract
We here reconstitute a minimal mammalian mitochondrial DNA (mtDNA) replisome in vitro. The mtDNA polymerase (POLgamma) cannot use double-stranded DNA (dsDNA) as template for DNA synthesis. Similarly, the TWINKLE DNA helicase is unable to unwind longer stretches of dsDNA. In combination, POLgamma and TWINKLE form a processive replication machinery, which can use dsDNA as template to synthesize single-stranded DNA (ssDNA) molecules of about 2 kb. The addition of the mitochondrial ssDNA-binding protein stimulates the reaction further, generating DNA products of about 16 kb, the size of the mammalian mtDNA molecule. The observed DNA synthesis rate is 180 base pairs (bp)/min, corresponding closely to the previously calculated value of 270 bp/min for in vivo DNA replication. Our findings provide the first biochemical evidence that TWINKLE is the helicase at the mitochondrial DNA replication fork. Furthermore, mutations in TWINKLE and POLgamma cause autosomal dominant progressive external ophthalmoplegia (adPEO), a disorder associated with deletions in mitochondrial DNA. The functional interactions between TWINKLE and POLgamma thus explain why mutations in these two proteins cause an identical syndrome.
Figures
Figure 1
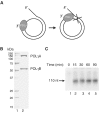
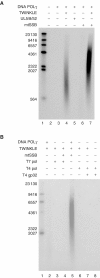
POLγ cannot use dsDNA as template for DNA synthesis. (A) The mini-circle template was prepared as described in Materials and methods. DNA synthesis is initiated at the 3′-hydroxy terminus and proceeds 20 nt before it encounters the dsDNA region of the template. (B) Recombinant POLγ (1 μg) purified over heparin sepharose was separated by SDS–PAGE (12.5%) and revealed with Coomassie brilliant blue staining. (C) POLγ (300 fmol) was incubated together with the mini-circle template at 37°C in a total reaction mixture of 75 μl as described in Materials and methods. At times indicated, 10 μl was removed and the reaction was terminated by addition of 10 μl of gel-loading buffer (98% formamide, 10 mM EDTA (pH 8.0), 0.025% xylene cyanol FF, and 0.025% bromophenol blue). The samples were analyzed on a 10% denaturing polyacrylamide gel.
Figure 2
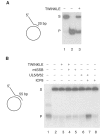
TWINKLE cannot unwind longer stretches of double-stranded DNA, even in the presence of mtSSB. (A) TWINKLE was added to a reaction mixture containing the short 20-bp double-stranded template as described in Materials and methods, and incubated for 30 min. Lane 1, untreated substrate; lane 2, substrate heated to 100°C before loading; lane 3, 550 fmol Twinkle; S, double-stranded substrate; P, single-stranded product. (B) TWINKLE is unable to unwind a 55-bp double-stranded template. Lane 3, 550 fmol Twinkle; lane 4, 550 fmol TWINKLE and 5 pmol mtSSB; lane 5, 5 pmol mtSSB; lane 6, 550 fmol UL5/52/8; lane 7, 550 fmol UL5/52/8 and 5 pmol ICP8; lane 8, 5 pmol ICP8.
Figure 3
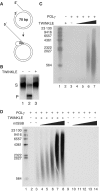
Rolling-circle DNA replication. (A) The mini-circle template was prepared as described in Materials and methods, and [α-32P]dCTP was used to preferentially label the leading strand. (B) TWINKLE alone cannot unwind the mini-circle template. TWINKLE was added to a reaction mixture containing the mini-circle template as described in Materials and methods, and incubated for 30 min. Lane 1, untreated substrate; lane 2, substrate heated to 100°C before loading; lane 3, 550 fmol TWINKLE; S, double-stranded substrate; P, single-stranded product. (C) Increasing amounts of TWINKLE were added to the mini-circle template together with 100 fmol POLγ as described under Materials and methods, and then analyzed on a 0.8% denaturating agarose gel. Lane 1, size marker; lane 2, 1.65 pmol TWINKLE (hexamer); lane 3, 0 fmol TWINKLE; lane 4, 55 fmol TWINKLE; lane 5, 180 fmol TWINKLE; lane 6, 550 fmol TWINKLE; lane 7, 1.65 pmol Twinkle. (D) A constant amount of TWINKLE (550 fmol, hexamer) and POLγ (100 fmol) was added when indicated, together with an increasing amount of mtSSB and analyzed as above. Lane 1, size marker; lane 3, 0 pmol mtSSB; lane 4, 0.1 pmol mtSSB; lane 5, 0.5 pmol mtSSB; lane 6, 1.0 pmol mtSSB; lane 7, 5 pmol mtSSB; lane 8, 10 pmol mtSSB; lane 9, 0 pmol mtSSB; lane 10, 0.1 pmol mtSSB; lane 11, 0.5 pmol mtSSB; lane 12, 1.0 pmol mtSSB; lane 13, 5 pmol mtSSB; lane 14, 10 pmol mtSSB.
Figure 4
The mitochondrial replication machinery can utilize both ATP and GTP as energy sources. TWINKLE (0.55 pmol), DNA POLγ (100 fmol), and mtSSB (5 pmol) were added to the reaction, together with 4 mM of the indicated NTP. In lane 2, we added 1 mM of each NTP. The reactions were incubated at 37°C for 60 min, and treated as described in Materials and methods. The reactions were analyzed on a 0.8% denaturating agarose gel.
Figure 5
(A) The UL5/8/52 DNA helicase cannot replace TWINKLE at the mtDNA replication fork. We added POLγ (100 fmol), TWINKLE (0.9 pmol), mtSSB (5 pmol), and UL5/52/8 (0.9 pmol). The reactions were incubated for 60 min at 37°C and treated as described under Materials and methods. (B) The T4 and T7 DNA polymerases cannot replace POLγ at the mtDNA replication fork. The replication reactions were as described in (A), with T4 pol (1 U), T7 (1 U), and T4 gp32 (5 pmol). Reactions were performed with 4 mM ATP, but identical results were obtained with GTP as cofactor (data not shown).
Figure 6
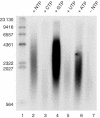
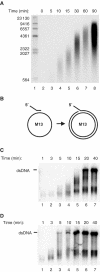
The mitochondrial DNA replication machinery can synthesize ssDNA at a rate of 180 bp/min on the dsDNA mini-circle template. (A) We incubated TWINKLE (2.7 pmol), DNA POLγ (500 fmol, mtSSB (25 pmol), and the mini-circle template (175 fmol) at 37°C in a total reaction volume of 125 μl as described in Materials and methods. At times indicated, we removed 12.5 μl and the reaction was stopped as described in Materials and methods. The reactions were analyzed on a 0.8% denaturating agarose gel. (B) A schematic representation of the single-stranded M13 mp18 DNA template used to measure DNA synthesis rate. (C) The DNA synthesis rate of POLγ on single-stranded M13 mp18 DNA in the presence of mtSSB was about 350 nt/min. DNA POLγ (500 fmol) and 25 pmol mtSSB were incubated together with a primed M13 ssDNA at 37°C in a total reaction mixture of 125 μl as described in Materials and methods. At times indicated, 12.5 μl was removed and the reaction was terminated by addition of 3-μl of stop solution. The reactions were analyzed by nondenaturing agarose gel electrophoresis. (D) The rate of T7 DNA polymerase together with mtSSB on ssDNA was 1500 nt/min. The reactions were as described in (C), but T7 DNA polymerase (5 U) was used instead of POLγ.
Similar articles
- The accessory subunit B of DNA polymerase gamma is required for mitochondrial replisome function.
Farge G, Pham XH, Holmlund T, Khorostov I, Falkenberg M. Farge G, et al. Nucleic Acids Res. 2007;35(3):902-11. doi: 10.1093/nar/gkl1116. Epub 2007 Jan 23. Nucleic Acids Res. 2007. PMID: 17251196 Free PMC article. - Progressive external ophthalmoplegia and multiple mitochondrial DNA deletions.
Van Goethem G, Martin JJ, Van Broeckhoven C. Van Goethem G, et al. Acta Neurol Belg. 2002 Mar;102(1):39-42. Acta Neurol Belg. 2002. PMID: 12094562 Review. - Subnormal levels of POLγA cause inefficient initiation of light-strand DNA synthesis and lead to mitochondrial DNA deletions and progressive external ophthalmoplegia [corrected].
Roos S, Macao B, Fusté JM, Lindberg C, Jemt E, Holme E, Moslemi AR, Oldfors A, Falkenberg M. Roos S, et al. Hum Mol Genet. 2013 Jun 15;22(12):2411-22. doi: 10.1093/hmg/ddt094. Epub 2013 Feb 27. Hum Mol Genet. 2013. PMID: 23446635 - TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication.
Milenkovic D, Matic S, Kühl I, Ruzzenente B, Freyer C, Jemt E, Park CB, Falkenberg M, Larsson NG. Milenkovic D, et al. Hum Mol Genet. 2013 May 15;22(10):1983-93. doi: 10.1093/hmg/ddt051. Epub 2013 Feb 7. Hum Mol Genet. 2013. PMID: 23393161 Free PMC article. - The adenine nucleotide translocase type 1 (ANT1): a new factor in mitochondrial disease.
Sharer JD. Sharer JD. IUBMB Life. 2005 Sep;57(9):607-14. doi: 10.1080/15216540500217735. IUBMB Life. 2005. PMID: 16203679 Review.
Cited by
- Biochemical Characterization of the Human Mitochondrial Replicative Twinkle Helicase: SUBSTRATE SPECIFICITY, DNA BRANCH MIGRATION, AND ABILITY TO OVERCOME BLOCKADES TO DNA UNWINDING.
Khan I, Crouch JD, Bharti SK, Sommers JA, Carney SM, Yakubovskaya E, Garcia-Diaz M, Trakselis MA, Brosh RM Jr. Khan I, et al. J Biol Chem. 2016 Jul 1;291(27):14324-14339. doi: 10.1074/jbc.M115.712026. Epub 2016 May 11. J Biol Chem. 2016. PMID: 27226550 Free PMC article. - mtDNA extramitochondrial replication mediates mitochondrial defect effects.
Shan Z, Li S, Gao Y, Jian C, Ti X, Zuo H, Wang Y, Zhao G, Wang Y, Zhang Q. Shan Z, et al. iScience. 2024 Jan 19;27(2):108970. doi: 10.1016/j.isci.2024.108970. eCollection 2024 Feb 16. iScience. 2024. PMID: 38322987 Free PMC article. - Human mitochondrial DNA helicase TWINKLE is both an unwinding and annealing helicase.
Sen D, Nandakumar D, Tang GQ, Patel SS. Sen D, et al. J Biol Chem. 2012 Apr 27;287(18):14545-56. doi: 10.1074/jbc.M111.309468. Epub 2012 Mar 1. J Biol Chem. 2012. PMID: 22383523 Free PMC article. - The Maintenance of Mitochondrial DNA Integrity and Dynamics by Mitochondrial Membranes.
Chapman J, Ng YS, Nicholls TJ. Chapman J, et al. Life (Basel). 2020 Aug 26;10(9):164. doi: 10.3390/life10090164. Life (Basel). 2020. PMID: 32858900 Free PMC article. Review. - Whole cell formaldehyde cross-linking simplifies purification of mitochondrial nucleoids and associated proteins involved in mitochondrial gene expression.
Rajala N, Hensen F, Wessels HJ, Ives D, Gloerich J, Spelbrink JN. Rajala N, et al. PLoS One. 2015 Feb 19;10(2):e0116726. doi: 10.1371/journal.pone.0116726. eCollection 2015. PLoS One. 2015. PMID: 25695250 Free PMC article.
References
- Ahnert P, Patel SS (1997) Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J Biol Chem 272: 32267–32273 - PubMed
- Benkovic SJ, Valentine AM, Salinas F (2001) Replisome-mediated DNA replication. Annu Rev Biochem 70: 181–208 - PubMed
- Carrodeguas JA, Kobayashi R, Lim SE, Copeland WC, Bogenhagen DF (1999) The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol Cell Biol 19: 4039–4046 - PMC - PubMed
- Carrodeguas JA, Theis K, Bogenhagen DF, Kisker C (2001) Crystal structure and deletion analysis show that the accessory subunit of mammalian DNA polymerase gamma, Pol gamma B, functions as a homodimer. Mol Cell 7: 43–54 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases