Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs - PubMed (original) (raw)
Comparative Study
. 2004 Jun;14(6):1107-18.
doi: 10.1101/gr.1774904.
Affiliations
- PMID: 15173116
- PMCID: PMC419789
- DOI: 10.1101/gr.1774904
Comparative Study
Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs
Haiyuan Yu et al. Genome Res. 2004 Jun.
Abstract
Proteins function mainly through interactions, especially with DNA and other proteins. While some large-scale interaction networks are now available for a number of model organisms, their experimental generation remains difficult. Consequently, interolog mapping--the transfer of interaction annotation from one organism to another using comparative genomics--is of significant value. Here we quantitatively assess the degree to which interologs can be reliably transferred between species as a function of the sequence similarity of the corresponding interacting proteins. Using interaction information from Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and Helicobacter pylori, we find that protein-protein interactions can be transferred when a pair of proteins has a joint sequence identity >80% or a joint E-value <10(-70). (These "joint" quantities are the geometric means of the identities or E-values for the two pairs of interacting proteins.) We generalize our interolog analysis to protein-DNA binding, finding such interactions are conserved at specific thresholds between 30% and 60% sequence identity depending on the protein family. Furthermore, we introduce the concept of a "regulog"--a conserved regulatory relationship between proteins across different species. We map interologs and regulogs from yeast to a number of genomes with limited experimental annotation (e.g., Arabidopsis thaliana) and make these available through an online database at http://interolog.gersteinlab.org. Specifically, we are able to transfer approximately 90,000 potential protein-protein interactions to the worm. We test a number of these in two-hybrid experiments and are able to verify 45 overlaps, which we show to be statistically significant.
Copyright 2004 Cold Spring Harbor Laboratory Press
Figures
Figure 1
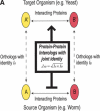
Schematic illustration of protein–protein interologs and the mapping methods. (A) Original interolog mapping. Theoretically, A-A′ and B-B′ should be orthologs between the two organisms. Operationally, only best-matching homologs are required. (B) Generalized interolog mapping. Proteins A1′, A2′, A3′, and A4′ in the target organism are all homologs of protein A in the source organism. These proteins form the A′ family. Likewise, protein B's homologs (B1′, B2′, B3′) form the B′ family in the target organism. If we know that protein A interacts with B, we can predict that the A′ family and the B′ family are interacting families. All possible pairs between these two families are considered as the generalized interologs (shown as black, dashed lines with arrows). (C) Comparison with the gold standards. After the interactions in the source organism are mapped onto the target organism, the predictions (i.e., generalized interologs) are compared with the gold standard positives and negatives. True positives are the predictions that overlap with the gold standard positives. False positives are those that overlap with the gold standard negatives. (D) Schematic illustration of protein–DNA interologs and regulogs. In the source organism, TF A binds to its binding site (SA) and regulates the downstream gene B. To perform the regulog mapping, TF A′ in the target organism needs to be the ortholog of A. Proteins B and B′ should also be orthologs. The DNA sequence upstream of gene B′ needs to contain the same motif (SA′) as SA. However, practically TF A and A′ only need to share ≥30% identity. The interaction between TF A′ and SA′ is the protein–DNA interolog of that between A and SA. The regulatory relationships between A → B and A′ → B′ are regulogs.
Figure 1
Schematic illustration of protein–protein interologs and the mapping methods. (A) Original interolog mapping. Theoretically, A-A′ and B-B′ should be orthologs between the two organisms. Operationally, only best-matching homologs are required. (B) Generalized interolog mapping. Proteins A1′, A2′, A3′, and A4′ in the target organism are all homologs of protein A in the source organism. These proteins form the A′ family. Likewise, protein B's homologs (B1′, B2′, B3′) form the B′ family in the target organism. If we know that protein A interacts with B, we can predict that the A′ family and the B′ family are interacting families. All possible pairs between these two families are considered as the generalized interologs (shown as black, dashed lines with arrows). (C) Comparison with the gold standards. After the interactions in the source organism are mapped onto the target organism, the predictions (i.e., generalized interologs) are compared with the gold standard positives and negatives. True positives are the predictions that overlap with the gold standard positives. False positives are those that overlap with the gold standard negatives. (D) Schematic illustration of protein–DNA interologs and regulogs. In the source organism, TF A binds to its binding site (SA) and regulates the downstream gene B. To perform the regulog mapping, TF A′ in the target organism needs to be the ortholog of A. Proteins B and B′ should also be orthologs. The DNA sequence upstream of gene B′ needs to contain the same motif (SA′) as SA. However, practically TF A and A′ only need to share ≥30% identity. The interaction between TF A′ and SA′ is the protein–DNA interolog of that between A and SA. The regulatory relationships between A → B and A′ → B′ are regulogs.
Figure 1
Schematic illustration of protein–protein interologs and the mapping methods. (A) Original interolog mapping. Theoretically, A-A′ and B-B′ should be orthologs between the two organisms. Operationally, only best-matching homologs are required. (B) Generalized interolog mapping. Proteins A1′, A2′, A3′, and A4′ in the target organism are all homologs of protein A in the source organism. These proteins form the A′ family. Likewise, protein B's homologs (B1′, B2′, B3′) form the B′ family in the target organism. If we know that protein A interacts with B, we can predict that the A′ family and the B′ family are interacting families. All possible pairs between these two families are considered as the generalized interologs (shown as black, dashed lines with arrows). (C) Comparison with the gold standards. After the interactions in the source organism are mapped onto the target organism, the predictions (i.e., generalized interologs) are compared with the gold standard positives and negatives. True positives are the predictions that overlap with the gold standard positives. False positives are those that overlap with the gold standard negatives. (D) Schematic illustration of protein–DNA interologs and regulogs. In the source organism, TF A binds to its binding site (SA) and regulates the downstream gene B. To perform the regulog mapping, TF A′ in the target organism needs to be the ortholog of A. Proteins B and B′ should also be orthologs. The DNA sequence upstream of gene B′ needs to contain the same motif (SA′) as SA. However, practically TF A and A′ only need to share ≥30% identity. The interaction between TF A′ and SA′ is the protein–DNA interolog of that between A and SA. The regulatory relationships between A → B and A′ → B′ are regulogs.
Figure 1
Schematic illustration of protein–protein interologs and the mapping methods. (A) Original interolog mapping. Theoretically, A-A′ and B-B′ should be orthologs between the two organisms. Operationally, only best-matching homologs are required. (B) Generalized interolog mapping. Proteins A1′, A2′, A3′, and A4′ in the target organism are all homologs of protein A in the source organism. These proteins form the A′ family. Likewise, protein B's homologs (B1′, B2′, B3′) form the B′ family in the target organism. If we know that protein A interacts with B, we can predict that the A′ family and the B′ family are interacting families. All possible pairs between these two families are considered as the generalized interologs (shown as black, dashed lines with arrows). (C) Comparison with the gold standards. After the interactions in the source organism are mapped onto the target organism, the predictions (i.e., generalized interologs) are compared with the gold standard positives and negatives. True positives are the predictions that overlap with the gold standard positives. False positives are those that overlap with the gold standard negatives. (D) Schematic illustration of protein–DNA interologs and regulogs. In the source organism, TF A binds to its binding site (SA) and regulates the downstream gene B. To perform the regulog mapping, TF A′ in the target organism needs to be the ortholog of A. Proteins B and B′ should also be orthologs. The DNA sequence upstream of gene B′ needs to contain the same motif (SA′) as SA. However, practically TF A and A′ only need to share ≥30% identity. The interaction between TF A′ and SA′ is the protein–DNA interolog of that between A and SA. The regulatory relationships between A → B and A′ → B′ are regulogs.
Figure 2
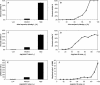
Conservation of protein–protein interactions between homologous protein pairs. (A,B) Relationships between V and J I. (C,D) Relationships between V and JE.(E,F) Relationships between L and J E. (A,C,E) Calculated based on the results from worm-yeast mapping. (B,D,F) The weighted average obtained when the interactions in all four organisms (i.e., S. cerevisiae, C. elegans, D. melanogaster, and H. pylori) were mapped onto yeast. (A) Low: JI ≤ 10%; Medium: 20% ≤ _J_I ≤ 30%; High: JI ≥ 40%. (C,D) Low: 10–40 ≤ J E ≤ 10–10; Medium: 10–100 ≤ J E ≤ 10–50; High: J E ≤ 10–110. Error bars represent 95% CI calculated by a resampling algorithm (see Supplemental material).
Figure 3
Distribution of the number of generalized interologs as a function of joint _E_-value (J E). The dashed line represents the number of all predictions above a given JE, that is, G(J). The solid line represents the number of true positives above a given JE, that is, TP.
Figure 4
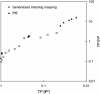
Comparison of generalized interolog mapping with PIE. In this figure, the plot (TP/|P′| versus TP/FP) is analogous to an ROC plot (TP/P vs. FP/N). Based on this curve, the performance of our method is comparable to that of the large-scale experimental data sets.
Figure 5
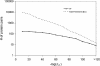
Conservation of protein–DNA interactions between homologous TFs. The conservation is measured as the relationships between V and I. The legend appears as an inset on the graph. The red, bold curve was calculated for all TFs in the source data sets (see Supplementary material). Error bars represent 95% CI calculated by the resampling algorithm.
Figure 6
Percentage of the overlaps between the predictions and different groups. (All) All experimentally determined interaction pairs; (Proteasome) interaction pairs involved in the 26S proteasome; (DDR) interaction pairs involved in DNA-damage repair; (Vulval-dev) interaction pairs involved in vulval development; (Others) interaction pairs involved in germ line, meiosis, metazoan, mitotic machinery, dauer formation, Chromosome III, chromatin remodeling, pharynx, and immunity. The _P_-values measuring the statistical significance of the overlaps between different groups and the predictions are given on top of each bar, which are calculated using the hypergeometric models (see Supplementary material).
Figure 7
Screenshot of the interolog/regulog database.
Similar articles
- Protein-protein interactions more conserved within species than across species.
Mika S, Rost B. Mika S, et al. PLoS Comput Biol. 2006 Jul 21;2(7):e79. doi: 10.1371/journal.pcbi.0020079. Epub 2006 May 18. PLoS Comput Biol. 2006. PMID: 16854211 Free PMC article. - 3D-interologs: an evolution database of physical protein- protein interactions across multiple genomes.
Lo YS, Chen YC, Yang JM. Lo YS, et al. BMC Genomics. 2010 Dec 1;11 Suppl 3(Suppl 3):S7. doi: 10.1186/1471-2164-11-S3-S7. BMC Genomics. 2010. PMID: 21143789 Free PMC article. - Filtering high-throughput protein-protein interaction data using a combination of genomic features.
Patil A, Nakamura H. Patil A, et al. BMC Bioinformatics. 2005 Apr 18;6:100. doi: 10.1186/1471-2105-6-100. BMC Bioinformatics. 2005. PMID: 15833142 Free PMC article. - Building a protein interaction map: research in the post-genome era.
Chen Z, Han M. Chen Z, et al. Bioessays. 2000 Jun;22(6):503-6. doi: 10.1002/(SICI)1521-1878(200006)22:6<503::AID-BIES2>3.0.CO;2-7. Bioessays. 2000. PMID: 10842303 Review. - Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey.
Promponas VJ, Ouzounis CA, Iliopoulos I. Promponas VJ, et al. Brief Bioinform. 2014 May;15(3):443-54. doi: 10.1093/bib/bbs072. Epub 2012 Dec 5. Brief Bioinform. 2014. PMID: 23220349 Free PMC article. Review.
Cited by
- Studying protein complexes by the yeast two-hybrid system.
Rajagopala SV, Sikorski P, Caufield JH, Tovchigrechko A, Uetz P. Rajagopala SV, et al. Methods. 2012 Dec;58(4):392-9. doi: 10.1016/j.ymeth.2012.07.015. Epub 2012 Jul 24. Methods. 2012. PMID: 22841565 Free PMC article. - SENSE-PPI reconstructs interactomes within, across, and between species at the genome scale.
Volzhenin K, Bittner L, Carbone A. Volzhenin K, et al. iScience. 2024 Jun 25;27(7):110371. doi: 10.1016/j.isci.2024.110371. eCollection 2024 Jul 19. iScience. 2024. PMID: 39055916 Free PMC article. - Computational prediction of protein-protein interactions in Leishmania predicted proteomes.
Rezende AM, Folador EL, Resende Dde M, Ruiz JC. Rezende AM, et al. PLoS One. 2012;7(12):e51304. doi: 10.1371/journal.pone.0051304. Epub 2012 Dec 10. PLoS One. 2012. PMID: 23251492 Free PMC article. - Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review.
Csermely P, Korcsmáros T, Kiss HJ, London G, Nussinov R. Csermely P, et al. Pharmacol Ther. 2013 Jun;138(3):333-408. doi: 10.1016/j.pharmthera.2013.01.016. Epub 2013 Feb 4. Pharmacol Ther. 2013. PMID: 23384594 Free PMC article. Review. - State of the art in silico tools for the study of signaling pathways in cancer.
Villaamil VM, Gallego GA, Cainzos IS, Valladares-Ayerbes M, Aparicio LMA. Villaamil VM, et al. Int J Mol Sci. 2012;13(6):6561-6581. doi: 10.3390/ijms13066561. Epub 2012 May 29. Int J Mol Sci. 2012. PMID: 22837650 Free PMC article. Review.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. - PubMed
- Andrade, M.A. and Sander, C. 1997. Bioinformatics: From genome data to biological knowledge. Curr. Opin. Biotechnol. 8: 675–683. - PubMed
WEB SITE REFERENCE
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases