Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes - PubMed (original) (raw)
Intragenomic heterogeneity and intergenomic recombination among haloarchaeal rRNA genes
Yan Boucher et al. J Bacteriol. 2004 Jun.
Abstract
More than one copy of rRNA operons, which code for both the small-subunit (SSU) and large-subunit (LSU) rRNA, are often found in prokaryotes. It is generally assumed that all rRNA operons within a single cell are almost identical. A notable exception is the extremely halophilic archaeal genus Haloarcula, most species of which are known to harbor highly divergent rRNA operons that differ at approximately 5% of the nucleotide positions in the SSU gene and at 1 to 2% of the nucleotide positions in the LSU gene. We report that such intragenomic heterogeneity is not unique to Haloarcula, as high levels of intragenomic sequence variation have been observed for the SSU genes of two other genera of extreme halophiles, Halosimplex and Natrinema. To investigate this in detail, the two rRNA operons of Halosimplex carlsbadense and the four operons of Natrinema sp. strain XA3-1 were cloned and completely sequenced. The SSU and LSU genes of H. carlsbadense show the highest levels of intragenomic heterogeneity observed so far in archaea (6.7 and 2.6%). The operons of Natrinema sp. strain XA3-1 have additional unusual characteristics, such as identical internal transcribed spacers, while one of four SSU genes is 5% divergent and all LSU genes differ from each other by 0.9 to 1.9%. The heterogeneity among the Natrinema sp. strain XA3-1 LSU genes is localized in hot spots, and one of these regions is shown to be the result of a recombination event with a distantly related halophile. This is the first example of interspecies recombination between rRNA genes in archaea, and the recombination occurred over one of the largest phylogenetic distances ever reported for such an event. We suggest that intragenomic heterogeneity of rRNA operons is an ancient and stable trait in several lineages of the Halobacteriales. The impact of this phenomenon on the taxonomy of extremely halophilic archaea is discussed.
Figures
FIG. 1.
Schematic diagram of the rRNA operon sequencing strategy. Cloned rRNA operons were sequenced by using primers matching regions conserved in all Halobacteriales SSU and LSU genes. For each operon, both DNA strands were sequenced by using one sequencing primer for every ∼500 bp on each strand. The ITS is the ITS between the SSU and LSU genes. Fragments HSIMP1 to HSIMP6 are the PCR fragments used to obtain the complete sequence of the H. carlsbadense second rRNA operon. The primers used to amplify these fragments are the sequencing primers matching the 5′ and 3′ ends.
FIG. 2.
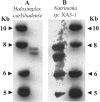
Southern hybridization of SSU gene probes to complete genomic DNA restriction endonuclease digests. The restriction endonucleases used were ClaI for Natrinema sp. strain XA3-1 (A) and NotI and ScaI for H. carlsbadense (B). The SSU gene probes used were directly amplified from genomic DNA of the species to which they were hybridized.
FIG.3.
Mapping of variable positions in the SSU (A) and LSU (B) genes based on the secondary structure of the rRNA product. Positions displaying intragenomic heterogeneity are indicated. Helices present in the secondary structure are numbered 1 to 45 (SSU gene) and 1 to 101 (LSU gene). Structural domains are indicated as follows: 5′, central (C), 3′M (major), and 3′m (minor) for the SSU gene and I to VI for the LSU gene. The relative evolutionary rate of each nucleotide position was calculated for the SSU gene in the Halobacteriales. The rate categories for the positions are indicated by colors in the secondary structure.
FIG. 4.
(A) Locations of the variable positions of Natrinema sp. strain XA3-1 LSU genes in the secondary structure of the rRNA product. Positions displaying intragenomic heterogeneity are indicated by gray triangles. The numbered boxes indicate different regions of the LSU genes in which heterogeneity between the multiple copies is observed, as follows: box 1, divergence of LSU-B at 27 positions between nucleotides 249 and 370; box 2, divergence of LSU-B and -C from LSU-A and -D at 13 positions between nucleotides 1113 and 1284; box 3, divergence of LSU-D at 13 positions between nucleotides 1523 and 1652; and box 4, divergence of LSU-C at 6 positions between nucleotides 1786 and 1856. Variable positions that are not in a box for the most part represent divergence in LSU-A. (B) Alignment of all available Halobacteriales LSU genes from positions 200 to 400, which corresponds to box 1 in the secondary structure diagram. At a given position, a nucleotide identical to the nucleotide in the Natrinema sp. strain XA3-1 LSU-A sequence is indicated by a dot.
FIG.5.
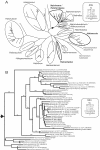
(A) Best maximum-likelihood distance tree for the SSU gene for the archaeal order Halobacteriales. (B) Best maximum-likelihood tree for the Halobacteriales subgroup in the triangular area in panel A. The evolutionary models and parameters used for the phylogenetic analyses are indicated in the boxes (NJ, neighbor joining; ML, maximum likelihood; GTR, general time reversible; I, proportion of invariable sites; G, gamma-distributed among-site rate variation; α, gamma rate shape parameter alpha; Γ, number of gamma distribution rate categories). The genera in which single organisms contain highly heterogeneous rRNA operons are indicated by boldface type. Bootstrap values were obtained by using a distance maximum-likelihood tree reconstruction method (only values greater than 50% are indicated at the nodes).
FIG. 6.
Comparison of best maximum-likelihood trees for the SSU and LSU genes of all genera of the Halobacteriales for which sequences of both genes were available. The evolutionary models and parameters used for the phylogenetic analyses are indicated (ML, maximum likelihood; GTR, general time reversible; I, proportion of invariable sites; α, gamma rate shape parameter alpha; Γ, number of gamma distribution rate categories). The numbers of nucleotide positions present in the edited alignment used in the phylogenetic analysis are also indicated. The bootstrap values at the nodes are the consensus values for maximum-likelihood trees for 1,000 pseudoreplicates.
Similar articles
- Evolution of the RNA polymerase B' subunit gene (rpoB') in Halobacteriales: a complementary molecular marker to the SSU rRNA gene.
Walsh DA, Bapteste E, Kamekura M, Doolittle WF. Walsh DA, et al. Mol Biol Evol. 2004 Dec;21(12):2340-51. doi: 10.1093/molbev/msh248. Epub 2004 Sep 8. Mol Biol Evol. 2004. PMID: 15356285 - Heterogeneous rRNAs are differentially expressed during the morphological development of Streptomyces coelicolor.
Kim HL, Shin EK, Kim HM, Ryou SM, Kim S, Cha CJ, Bae J, Lee K. Kim HL, et al. FEMS Microbiol Lett. 2007 Oct;275(1):146-52. doi: 10.1111/j.1574-6968.2007.00872.x. Epub 2007 Aug 15. FEMS Microbiol Lett. 2007. PMID: 17711457 - Intragenomic heterogeneity and intergenomic recombination among Vibrio parahaemolyticus 16S rRNA genes.
Harth E, Romero J, Torres R, Espejo RT. Harth E, et al. Microbiology (Reading). 2007 Aug;153(Pt 8):2640-2647. doi: 10.1099/mic.0.2007/009175-0. Microbiology (Reading). 2007. PMID: 17660428 - Taxonomy of halophilic Archaea: current status and future challenges.
Oren A. Oren A. Extremophiles. 2014 Sep;18(5):825-34. doi: 10.1007/s00792-014-0654-9. Epub 2014 Aug 8. Extremophiles. 2014. PMID: 25102811 Review. - Intragenomic mutational heterogeneity: structural and functional insights from gene evolution.
Hara Y, Kuraku S. Hara Y, et al. Trends Genet. 2025 May 5:S0168-9525(25)00075-7. doi: 10.1016/j.tig.2025.03.007. Online ahead of print. Trends Genet. 2025. PMID: 40328580 Review.
Cited by
- Expression of divergent LSU rRNA genes in the Vibrio vulnificus CMCP6 genome during both infection and non-pathogenic stages.
Kim HL, Ryou SM, Lee M, Lee JW, Lee K, Bae J. Kim HL, et al. Curr Microbiol. 2011 Jan;62(1):133-8. doi: 10.1007/s00284-010-9684-4. Epub 2010 Jun 5. Curr Microbiol. 2011. PMID: 20526601 - Analysis of intergenic spacer region length polymorphisms to investigate the halophilic archaeal diversity of stromatolites and microbial mats.
Leuko S, Goh F, Allen MA, Burns BP, Walter MR, Neilan BA. Leuko S, et al. Extremophiles. 2007 Jan;11(1):203-10. doi: 10.1007/s00792-006-0028-z. Epub 2006 Nov 3. Extremophiles. 2007. PMID: 17082971 - Haloglomus irregulare gen. nov., sp. nov., a New Halophilic Archaeon Isolated from a Marine Saltern.
Durán-Viseras A, Sánchez-Porro C, Ventosa A. Durán-Viseras A, et al. Microorganisms. 2020 Feb 2;8(2):206. doi: 10.3390/microorganisms8020206. Microorganisms. 2020. PMID: 32024278 Free PMC article. - Evidence from phylogenetic and genome fingerprinting analyses suggests rapidly changing variation in Halorubrum and Haloarcula populations.
Ram Mohan N, Fullmer MS, Makkay AM, Wheeler R, Ventosa A, Naor A, Gogarten JP, Papke RT. Ram Mohan N, et al. Front Microbiol. 2014 Apr 9;5:143. doi: 10.3389/fmicb.2014.00143. eCollection 2014. Front Microbiol. 2014. PMID: 24782838 Free PMC article. - Expression and Function of Different Guanine-Plus-Cytosine Content 16S rRNA Genes in Haloarcula hispanica at Different Temperatures.
Sato Y, Fujiwara T, Kimura H. Sato Y, et al. Front Microbiol. 2017 Mar 28;8:482. doi: 10.3389/fmicb.2017.00482. eCollection 2017. Front Microbiol. 2017. PMID: 28400752 Free PMC article.
References
- Amann, G., K. O. Stetter, E. Llobet-Brossa, R. Amann, and J. Anton. 2000. Direct proof for the presence and expression of two 5% different 16S rRNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373-376. - PubMed
- Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. - PubMed
- Carranza, S., G. Giribet, C. Ribera, Baguna, and M. Riutort. 1996. Evidence that two types of 18S rDNA coexist in the genome of Dugesia (Schmidtea) mediterranea (Platyhelminthes, Turbellaria, Tricladida). Mol. Biol. Evol. 13:824-832. - PubMed
- Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol. 13:451-461. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases