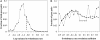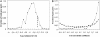Coevolution of gene expression among interacting proteins - PubMed (original) (raw)
Coevolution of gene expression among interacting proteins
Hunter B Fraser et al. Proc Natl Acad Sci U S A. 2004.
Abstract
Physically interacting proteins or parts of proteins are expected to evolve in a coordinated manner that preserves proper interactions. Such coevolution at the amino acid-sequence level is well documented and has been used to predict interacting proteins, domains, and amino acids. Interacting proteins are also often precisely coexpressed with one another, presumably to maintain proper stoichiometry among interacting components. Here, we show that the expression levels of physically interacting proteins coevolve. We estimate average expression levels of genes from four closely related fungi of the genus Saccharomyces using the codon adaptation index and show that expression levels of interacting proteins exhibit coordinated changes in these different species. We find that this coevolution of expression is a more powerful predictor of physical interaction than is coevolution of amino acid sequence. These results demonstrate that gene expression levels can coevolve, adding another dimension to the study of the coevolution of interacting proteins and underscoring the importance of maintaining coexpression of interacting proteins over evolutionary time. Our results also suggest that expression coevolution can be used for computational prediction of protein-protein interactions.
Figures
Fig. 1.
Coevolution of sequence. (A) A histogram of the base 10 logarithms of variance in evolutionary rates for all 192,510 possible pairs of proteins in this study. The variance for each protein in a pair was calculated, and the lower of the two was used to represent the pair. The dashed line indicates the variance cutoff described in the main text. Note that evolutionary rates were normalized by the mean rate for each branch of the phylogenetic tree (see Materials and Methods). (B) A histogram of the correlation coefficients indicating the strength of amino acid sequence coevolution for 200 pairs of interacting proteins (solid line) and 26,596 pairs of noninteracting proteins (dashed line). The two distributions are significantly different from one another (KS test, P = 0.0069). Bin labels are the upper bound for each bin (e.g., the label 0.9 indicates 0.8 < r ≤ 0.9).
Fig. 2.
Coevolution of expression. (A) A histogram of the base 10 logarithms of variance in CAI for all 192,510 possible pairs of the 621 proteins in this study. The variance for each protein in a pair was calculated, and the lower of the two was used to represent the pair. The dashed line indicates the variance cutoff described in the main text. (B) A histogram of the correlation coefficients indicating the strength of CAI coevolution for 200 pairs of interacting proteins (solid line) and 11,581 pairs of noninteracting proteins (dashed line). The two distributions are significantly different from one another (KS test, P < 10-26). Bin labels are the upper bound for each bin (e.g., the label 0.9 indicates 0.8 < r ≤ 0.9).
Similar articles
- Coevolution is a short-distance force at the protein interaction level and correlates with the modular organization of protein networks.
Liang Z, Xu M, Teng M, Niu L, Wu J. Liang Z, et al. FEBS Lett. 2010 Oct 8;584(19):4237-40. doi: 10.1016/j.febslet.2010.09.014. Epub 2010 Sep 17. FEBS Lett. 2010. PMID: 20837013 - Correlation and prediction of gene expression level from amino acid and dipeptide composition of its protein.
Raghava GP, Han JH. Raghava GP, et al. BMC Bioinformatics. 2005 Mar 17;6:59. doi: 10.1186/1471-2105-6-59. BMC Bioinformatics. 2005. PMID: 15773999 Free PMC article. - Multifunctionality dominantly determines the rate of human housekeeping and tissue specific interacting protein evolution.
Podder S, Mukhopadhyay P, Ghosh TC. Podder S, et al. Gene. 2009 Jun 15;439(1-2):11-6. doi: 10.1016/j.gene.2009.03.005. Epub 2009 Mar 20. Gene. 2009. PMID: 19306918 - An integrated view of molecular coevolution in protein-protein interactions.
Lovell SC, Robertson DL. Lovell SC, et al. Mol Biol Evol. 2010 Nov;27(11):2567-75. doi: 10.1093/molbev/msq144. Epub 2010 Jun 14. Mol Biol Evol. 2010. PMID: 20551042 Review. - Synonymous codon usage in Saccharomyces cerevisiae.
Sharp PM, Cowe E. Sharp PM, et al. Yeast. 1991 Oct;7(7):657-78. doi: 10.1002/yea.320070702. Yeast. 1991. PMID: 1776357 Review. No abstract available.
Cited by
- Chapter 4: Protein interactions and disease.
Gonzalez MW, Kann MG. Gonzalez MW, et al. PLoS Comput Biol. 2012;8(12):e1002819. doi: 10.1371/journal.pcbi.1002819. Epub 2012 Dec 27. PLoS Comput Biol. 2012. PMID: 23300410 Free PMC article. - A conserved mammalian protein interaction network.
Pérez-Bercoff Å, Hudson CM, Conant GC. Pérez-Bercoff Å, et al. PLoS One. 2013;8(1):e52581. doi: 10.1371/journal.pone.0052581. Epub 2013 Jan 2. PLoS One. 2013. PMID: 23320073 Free PMC article. - Salmonid genomes have a remarkably expanded akirin family, coexpressed with genes from conserved pathways governing skeletal muscle growth and catabolism.
Macqueen DJ, Kristjánsson BK, Johnston IA. Macqueen DJ, et al. Physiol Genomics. 2010 Jun;42(1):134-48. doi: 10.1152/physiolgenomics.00045.2010. Epub 2010 Apr 13. Physiol Genomics. 2010. PMID: 20388840 Free PMC article. - Benchmarking ortholog identification methods using functional genomics data.
Hulsen T, Huynen MA, de Vlieg J, Groenen PM. Hulsen T, et al. Genome Biol. 2006;7(4):R31. doi: 10.1186/gb-2006-7-4-r31. Epub 2006 Apr 13. Genome Biol. 2006. PMID: 16613613 Free PMC article. - Specificity in protein interactions and its relationship with sequence diversity and coevolution.
Hakes L, Lovell SC, Oliver SG, Robertson DL. Hakes L, et al. Proc Natl Acad Sci U S A. 2007 May 8;104(19):7999-8004. doi: 10.1073/pnas.0609962104. Epub 2007 Apr 27. Proc Natl Acad Sci U S A. 2007. PMID: 17468399 Free PMC article.
References
- Altschuh, D., Lesk, A. M., Bloomer, A. C. & Klug, A. (1987) J. Mol. Biol. 193, 693-707. - PubMed
- Moyle, W. R., Campbell, R. K., Myers, R. V., Bernard, M. P., Han, Y. & Wang, X. (1994) Science 368, 251-255. - PubMed
- Pazos, F., Helmer-Citterich, M., Ausiello, G. & Valencia, A. (1997) J. Mol. Biol. 271, 511-523. - PubMed
- Goh, C. S., Bogan, A. A., Joachimiak, M., Walther, D. & Cohen, F. E. (2000) J. Mol. Biol. 299, 283-293. - PubMed
- Ramani, A. K. & Marcotte, E. M. (2003) J. Mol. Biol. 327, 273-284. - PubMed