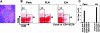CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion - PubMed (original) (raw)
CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion
Ann E Herman et al. J Exp Med. 2004.
Abstract
CD4+CD25+ T regulatory cells (Tregs) prevent autoimmune disease, yet little is known about precisely where they exert their influence naturally in a spontaneous autoimmune disorder. Here, we report that Tregs and T effector cells (Teffs) coexist within the pancreatic lesion before type 1 diabetes onset. We find that BDC2.5 T cell receptor transgenic animals contain a small subset of FoxP3 positive CD4+CD25+CD69- cells in the pancreas, actively turning over, expressing the clonotypic receptor, and containing functional regulatory activity. Gene expression profiling confirms that the CD4+CD25+CD69- cells in pancreatic tissue express transcripts diagnostic of regulatory cells, but with significantly higher levels of interleukin 10 and inducible costimulator (ICOS) than their lymph node counterparts. Blockade of ICOS rapidly converts early insulitis to diabetes, which disrupts the balance of Teffs and Tregs and promotes a very broad shift in the expression of the T regulatory-specific profile. Thus, CD4+CD25+69- Tregs operate directly in the autoimmune lesion and are dependent on ICOS to keep it in a nondestructive state.
Figures
Figure 1.
Infiltrate in the pancreatic lesion contains subsets of CD4+ cells with activated or regulatory phenotypes. (A) Islets of BDC2.5 mice at 4 wk of age. (B) Cells from pancreatic infiltrate of 3–4-wk-old BDC2.5 mice were isolated as aforementioned and analyzed for CD4, B220, live cell dye, CD25, and CD69 markers. Plots are gated on CD4+B220− live lymphocytes, and expression of activation markers CD25 and CD69 is shown. Data represent 10 separate experiments. (C) Pancreatic cells gated for CD4+ expression, live lymphocytes, and B220− cells were sorted (whole CD4+) or sorted separately into the distinct CD25/CD69 subsets observed in B. Whole BDC2.5 LNs, or LN sorted into CD4+CD25+ and CD4+CD25− subsets were also prepared. cDNA was prepared from each indicated group and assessed for expression of FoxP3 cDNA by real-time PCR analysis. Data are presented as relative expression of FoxP3 compared with the value in whole LNs (set to 1). Averaged results of three experiments are shown.
Figure 2.
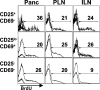
CD4+CD25+CD69−, CD25loCD69+, and CD25−CD69− subsets are proliferating in the pancreas and LNs of unmanipulated BDC2.5 mice. 3–4-wk-old BDC2.5 mice were fed BrdU in their drinking water for 5 d. Pancreas (Panc), pancreatic LNs (PLN), or inguinal LN (ILN) were isolated and analyzed by five-color flow cytometry for CD4, B220, CD25, CD69, and BrdU expression. Cells were gated for lymphocytes, CD4+B220− cells, and indicated CD25/CD69 subsets (as in Fig. 1 B). Isotype control (thick line) versus BrdU (thin line) staining is shown. Data are representative of three experiments.
Figure 3.
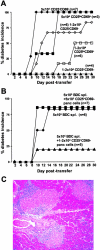
CD4+CD25+CD69− cells isolated directly from the pancreatic lesion protect from diabetes, whereas CD4+CD25loCD69+ and CD25−CD69− cells are pathogenic. (A) Cells from the pancreatic lesion of 3–4-wk-old BDC2.5 mice were sorted into CD4+CD25+CD69− (▴CD25+CD69−), CD4+CD25loCD69+ (▪CD25loCD69+), and CD4+CD25−CD69−(♦CD25−CD69−) subsets by flow cytometry. Sorted cells were transferred i.p. separately into NOD.scid recipients at two doses as follows: 50,000 cells (closed symbols) or 10–20,000 cells (open symbols). Mice were monitored every other day after 7 d for the onset of hyperglycemia by urine, and diabetes was confirmed by blood glucose levels >250 mg/dl. Data represent pooled results of three separate experiments with similar results. (B) CD4+CD25+CD69− cells were sorted as in A. Either 10–20,000 (▴), 5,000 (▪), or no (♦) CD4+CD25+CD69− cells were transferred i.p. into NOD.scid recipient animals in combination with 5 × 105 diabetogenic BDC2.5 splenocytes. Mice were monitored as described in A. Data represent pooled results of three separate experiments with similar results. (C) Histological sections were prepared from pancreata of transferred NOD.scid animals. Hematoxylin and eosin–stained sections from recipients that did not develop diabetes (protected by Treg) by the end of 30 d after receiving the 10–20,000 cell dose of CD4+CD25+CD69− cells and 5 × 105 diabetogenic BDC2.5 splenocytes are shown.
Figure 4.
CD4+CD25+CD69− cells have a gene expression profile of Tregs, whereas CD25loCD69+ and CD25−CD69− cells are distinct. (A) Cells were sorted to high purity directly from the pancreas lesion of 3–4-wk-old BDC2.5 mice for the CD4+CD25+CD69− population, or the CD25loCD69+ and CD25−CD69− combined populations. RNA was prepared and hybridized to Affymetrix U74Av2 array GeneChips® as described in Materials and Methods. RMA analysis was used to compare relative expression of data from the CD4+CD25+CD69− population to the CD25loCD69+ and CD25−CD69− combined populations. The cutoff for significant differences was determined using a random dataset to identify a 10% FPR for the real data, in this case a 2.1-fold increase (or decrease) in gene expression (highlighted in blue). For each cell type, three to five separate experiments were performed. (B) The gene list of 77 from McHugh et al. (reference 45) is highlighted on our relative gene expression graph in blue (up in LN CD25+) and red (up in LN CD25−).
Figure 5.
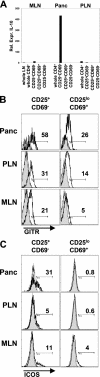
IL-10, GITR, and ICOS are specifically up-regulated in the pancreatic lesion compared with the draining lymph node. (A) Cells were sorted to high purity from the pancreas lesion, pancreatic LNs (PLN), or mesenteric LNs (MLN) of 3–4-wk-old BDC2.5 mice for the CD4+CD25+CD69−, CD25loCD69+, or CD25−CD69− populations. mRNA was prepared from each indicated group and assessed for IL-10 expression by real-time PCR analysis. Data are presented as relative expression of IL-10 compared with the value in whole LNs (set to 1). The average of three experiments is shown. (B) GITR expression (unshaded histograms) was examined on CD4+B220− live lymphocytes gated on the CD25/CD69 profiles indicated (Fig. 1 B) from the pancreas, PLN, or MLN and compared with isotype control (shaded histograms). The data shown are representative of three experiments. (C) ICOS expression (unshaded histograms) was examined on subsets as in B. The data shown are representative of five experiments.
Figure 6.
ICOS or CTLA-4 blockade in young BDC2.5 animals leads to rapid diabetes onset and correlates with a disruption of the T regulatory/T effector balance. (A) BDC2.5 mice were treated at 9 and 12 d of age with anti–CTLA-4 or ICOS mAb, indicated by arrow (left). Mice were treated with two doses of anti-ICOS mAb starting at 21, 26, or 35 d of age (arrows); with anti–CTLA-4 mAb starting at day 26; or with controls as described in Materials and Methods (right). Diabetes was monitored as aforementioned. (B) BDC2.5 mice were treated at 21 and 24 d with anti-ICOS mAb or control and killed at 26 d of age before diabetes onset. Pancreatic lesion, PLN, and ILN cells were prepared and stained with anti-CD4, B220, CD25, CD69, and live cell dye. Percentages are shown of pancreatic CD4+CD25loCD69+ cells (CD25loCD69+) or CD4+CD25+CD69− cells (CD25+CD69−), gated on live lymphocytes and B220− cells (n = 5; P < 0.008, CD25loCD69+ cells; P < 0.008, CD25+CD69− cells by pairwise Student's t test). PLN and ILN are not shown because there were no significant differences.
Figure 7.
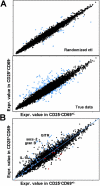
ICOS blockade dampens the immunoregulatory gene profile and correlates with expression of proinflammatory cytokines. (A) Cells from pancreatic infiltrate of 3–4-wk-old BDC2.5 mice were isolated after treatment with anti-ICOS mAb or rat IgG control as indicated, and analyzed for CD25/CD69 expression profiles on CD4+B220− live lymphocytes. Data represent five separate experiments. Cells were sorted to high purity directly from the pancreatic lesion of 3–4-wk-old BDC2.5 mice 5 d after indicated treatments for the CD4+CD25+CD69− Treg population, or the CD25loCD69+ and CD25−CD69− combined Teff populations. RNA was prepared and hybridized to Affymetrix U74Av2 array GeneChips®, and data were analyzed as described in Materials and Methods and in Fig. 4. (B) Expression value comparison plot for Teff populations with anti-ICOS versus control treatment. (C) Similar plot for Treg populations. (D) A ratio plot comparing the ratio of Treg anti-ICOS–treated/Teff anti-ICOS–treated profiles to the same ratio in the Treg and Teff control-treated profiles. Diagonal line indicates where points would fall if nothing changed between the treatments. A selection of genes that are highly over- or underexpressed after anti-ICOS treatment are highlighted. (E) Changes in expression values for cytokines within the lesion in Treg and Teff cells from each treatment group are shown. Asterisks indicate fold changes >1.9.
Similar articles
- Tolerogenic dendritic cells induce CD4+CD25hiFoxp3+ regulatory T cell differentiation from CD4+CD25-/loFoxp3- effector T cells.
Huang H, Dawicki W, Zhang X, Town J, Gordon JR. Huang H, et al. J Immunol. 2010 Nov 1;185(9):5003-10. doi: 10.4049/jimmunol.0903446. Epub 2010 Sep 24. J Immunol. 2010. PMID: 20870943 - Th1-Like ICOS+ Foxp3+ Treg Cells Preferentially Express CXCR3 and Home to β-Islets during Pre-Diabetes in BDC2.5 NOD Mice.
Kornete M, Mason ES, Girouard J, Lafferty EI, Qureshi S, Piccirillo CA. Kornete M, et al. PLoS One. 2015 May 6;10(5):e0126311. doi: 10.1371/journal.pone.0126311. eCollection 2015. PLoS One. 2015. PMID: 25946021 Free PMC article. - Costimulation via glucocorticoid-induced TNF receptor in both conventional and CD25+ regulatory CD4+ T cells.
Kanamaru F, Youngnak P, Hashiguchi M, Nishioka T, Takahashi T, Sakaguchi S, Ishikawa I, Azuma M. Kanamaru F, et al. J Immunol. 2004 Jun 15;172(12):7306-14. doi: 10.4049/jimmunol.172.12.7306. J Immunol. 2004. PMID: 15187106 - Control of type 1 autoimmune diabetes by naturally occurring CD4+CD25+ regulatory T lymphocytes in neonatal NOD mice.
Piccirillo CA, Tritt M, Sgouroudis E, Albanese A, Pyzik M, Hay V. Piccirillo CA, et al. Ann N Y Acad Sci. 2005 Jun;1051:72-87. doi: 10.1196/annals.1361.048. Ann N Y Acad Sci. 2005. PMID: 16126946 Review. - Dendritic cells expand antigen-specific Foxp3+ CD25+ CD4+ regulatory T cells including suppressors of alloreactivity.
Yamazaki S, Inaba K, Tarbell KV, Steinman RM. Yamazaki S, et al. Immunol Rev. 2006 Aug;212:314-29. doi: 10.1111/j.0105-2896.2006.00422.x. Immunol Rev. 2006. PMID: 16903923 Review.
Cited by
- CD4+CD126low/- Foxp3+ Cell Population Represents a Superior Subset of Regulatory T Cells in Treating Autoimmune Diseases.
Chen Y, Xu Z, Liang R, Wang J, Xu A, Na N, Li B, Wang R, Joseph M, Olsen N, Hsueh W, Zheng SG. Chen Y, et al. Mol Ther. 2020 Nov 4;28(11):2406-2416. doi: 10.1016/j.ymthe.2020.07.020. Epub 2020 Jul 21. Mol Ther. 2020. PMID: 32738192 Free PMC article. - The Role of FGL2 in the Pathogenesis and Treatment of Hepatitis C Virus Infection.
Shalev I, Selzner N, Helmy A, Foerster K, Adeyi OA, Grant DR, Levy G. Shalev I, et al. Rambam Maimonides Med J. 2010 Jul 2;1(1):e0004. doi: 10.5041/RMMJ.10004. Print 2010 Jul. Rambam Maimonides Med J. 2010. PMID: 23908776 Free PMC article. - Regulatory T Cell Numbers in Inflamed Skin Are Controlled by Local Inflammatory Cues That Upregulate CD25 and Facilitate Antigen-Driven Local Proliferation.
Billroth-MacLurg AC, Ford J, Rosenberg A, Miller J, Fowell DJ. Billroth-MacLurg AC, et al. J Immunol. 2016 Sep 15;197(6):2208-18. doi: 10.4049/jimmunol.1502575. Epub 2016 Aug 10. J Immunol. 2016. PMID: 27511734 Free PMC article. - Role of dendritic cell maturity/costimulation for generation, homeostasis, and suppressive activity of regulatory T cells.
Pletinckx K, Döhler A, Pavlovic V, Lutz MB. Pletinckx K, et al. Front Immunol. 2011 Sep 27;2:39. doi: 10.3389/fimmu.2011.00039. eCollection 2011. Front Immunol. 2011. PMID: 22566829 Free PMC article. - IL-10 and ICOS Differentially Regulate T Cell Responses in the Brain during Chronic Toxoplasma gondii Infection.
O'Brien CA, Batista SJ, Still KM, Harris TH. O'Brien CA, et al. J Immunol. 2019 Mar 15;202(6):1755-1766. doi: 10.4049/jimmunol.1801229. Epub 2019 Feb 4. J Immunol. 2019. PMID: 30718297 Free PMC article.
References
- Tisch, R., and H. McDevitt. 1996. Insulin-dependent diabetes mellitus. Cell. 85:291–297. - PubMed
- Lieberman, S.M., A.M. Evans, B. Han, T. Takaki, Y. Vinnitskaya, J.A. Caldwell, D.V. Serreze, J. Shabanowitz, D.F. Hunt, S.G. Nathenson, et al. 2003. Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 100:8384–8388. - PMC - PubMed
- Katz, J.D., B. Wang, K. Haskins, C. Benoist, and D. Mathis. 1993. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 74:1089–1100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- Z P30 DK36836-17/DK/NIDDK NIH HHS/United States
- R01 CA084500/CA/NCI NIH HHS/United States
- AI 39671/AI/NIAID NIH HHS/United States
- CA 84500/CA/NCI NIH HHS/United States
- T32 CA070083/CA/NCI NIH HHS/United States
- 2 T32 DK07260-25/DK/NIDDK NIH HHS/United States
- T32 DK007260/DK/NIDDK NIH HHS/United States
- P01 AI039671/AI/NIAID NIH HHS/United States
- T32 CA 70083-07/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous