Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway - PubMed (original) (raw)
Comparative Study
Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway
Pankaj Kapahi et al. Curr Biol. 2004.
Abstract
In many species, reducing nutrient intake without causing malnutrition extends lifespan. Like DR (dietary restriction), modulation of genes in the insulin-signaling pathway, known to alter nutrient sensing, has been shown to extend lifespan in various species. In Drosophila, the target of rapamycin (TOR) and the insulin pathways have emerged as major regulators of growth and size. Hence we examined the role of TOR pathway genes in regulating lifespan by using Drosophila. We show that inhibition of TOR signaling pathway by alteration of the expression of genes in this nutrient-sensing pathway, which is conserved from yeast to human, extends lifespan in a manner that may overlap with known effects of dietary restriction on longevity. In Drosophila, TSC1 and TSC2 (tuberous sclerosis complex genes 1 and 2) act together to inhibit TOR (target of rapamycin), which mediates a signaling pathway that couples amino acid availability to S6 kinase, translation initiation, and growth. We find that overexpression of dTsc1, dTsc2, or dominant-negative forms of dTOR or dS6K all cause lifespan extension. Modulation of expression in the fat is sufficient for the lifespan-extension effects. The lifespan extensions are dependent on nutritional condition, suggesting a possible link between the TOR pathway and dietary restriction.
Figures
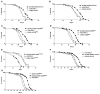
Figure 1. Regulation of Lifespan by Modulating Genes in the TOR Signaling Pathway
Survival curves on standard laboratory food for male flies obtained after crossing the ubiquitously expressed _da_-GAL4 enhancer trap with various UAS constructs carrying genes in the TOR signaling pathway ([A]–[E] at 29°C, [F] and [G] at 25°C). All logrank test p values are based on comparison of the overexpressor strain with the driver alone. The plus symbol indicates the parental strain w1118. The mean lifespan (days) of the various genotypes were as follows. (A) _da_-GAL4/UAS-dTsc1 (closed squares), 29 days (n=136); +/_da_-GAL4 (driver alone) (open triangles), 26 days (n = 92); +/UAS-dTsc1 (open circles), 25 days (n = 109) (p < 0.0001). (B) _da_-GAL4/UAS-dTsc2 (closed squares), 29 days (n = 112); +/_da_-GAL4 (open triangles), 26 days (n = 92); +/UAS-dTsc2 (open circles), 25 days (n = 118) (p < 0.0001). (C) _da_-GAL4/UAS-dTORFRB (closed squares), 30 days (n = 126); +/_da_-GAL4 (open triangles), 26 days (n = 92); +/UAS-dTORFRB (open circles), 22 days (n = 98) (p < 0.0001). (D) _da_-GAL4/UAS-dS6kKQ (closed squares), 30 days (n = 122); +/_da_-GAL4 (open triangles), 26 days (n = 92); +/UAS-dS6kKQ (open circles), 23 days (n = 115) (p < 0.0001). (E) _da_-GAL4/UAS-dS6kSTDETE (closed squares), 16 days (n = 75); +/_da_-GAL4 (open triangles), 26 days (n = 92); +/UAS-dS6kSTDETE (open circles), 24 days (n = 40) (p < 0.0001). (F) _da_-GAL4/UAS-dTsc2 (closed squares), 69 days (n = 134); +/_da_-GAL4 (open triangles), 58 days (n = 133); +/UAS-dTsc2 (open circles), 56 days (n = 126) (p < 0.0001). (G) _da_-GAL4/UAS-dTORFRB (closed squares), 72 days (n = 112); +/_da_-GAL4 (open triangles), 58 days (n = 133); +/UAS-dTORFRB (open circles), 55 days (n = 128) (p < 0.0001).
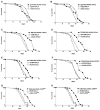
Figure 2. Dependence of Lifespan Extension on Overexpression in Specific Tissues
Survival curves, on standard laboratory food, for male flies obtained by crossing various genes in the TOR pathway by using different tissue specific drivers along with appropriate control strains. _gmr_-GAL4 is expressed in the eye, _appl_-GAL4 in the nervous system, 24B-GAL4 and PO188-GAL4 in the muscle and fat tissues, and DJ634-GAL4 and PO163-GAL4 in the fat tissue. All logrank test p values were based on comparison of the overexpressor strain with the driver alone. The plus symbol indicates the parental strain w1118. The mean lifespan at 29°C of the various genotypes were as follows. (A) _gmr_-GAL4/ UAS-dTsc2 (closed squares), 27 days (n = 69); +/_gmr_-GAL4 (open triangles), 31 days (n = 102); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.01). (B) _appl_-GAL4/ UAS-dTsc2 (closed squares), 33 days (n = 108); +/_appl_-GAL4 (open triangles), 32 days (n = 85); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.001). (C) 24B- GAL4/UAS-dTsc2 (closed squares), 37 days (n = 107); +/24B-GAL4 (open triangles), 31 days (n = 67); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (D) PO188-GAL4/UAS-dTsc2 (closed squares), 35 days (n = 76); +/PO188-GAL4 (open triangles), 24 days (n = 71); +/ UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (E) DJ634-GAL4/UAS-dTsc2 (closed squares), 30.6 days (n = 120); +/DJ634-GAL4 (open triangles), 24.6 days (n = 120); +/UAS-dTsc2 (open circles), 25.3 days (n = 118) (p < 0.0001). (F) PO163-GAL4/UAS-dTsc2 (closed squares), 36 days (n = 126); +/PO163-GAL4 (open triangles), 28 days (n = 87); +/UAS-dTsc2 (open circles), 27 days (n = 81) (p < 0.0001). (G) DJ634-GAL4/UAS-dTORFRB (closed squares), 31 days (n = 107); +/DJ634-GAL4 (open triangles), 25 days (n = 120); +/UAS-dTORFRB (open circles), 22 days (n = 98) (p < 0.0001). (H) DJ634-GAL4/UAS-dS6kKQ (closed squares), 31 days (n = 110); +/DJ634-GAL4 (open triangles), 25 days (n = 120); +/UAS-dS6kKQ (open circles), 23 days (n = 115) (p < 0.0001).
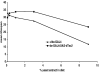
Figure 3. Lifespan Extension by dTsc2 Overexpression Is Dependent on the Concentration of Yeast Extract in the Diet
Logrank test was used to calculate the p values. The plus symbol indicates the parental strain w1118. Mean lifespan at 29°C of adult male _da_-Gal4/+ (closed triangles) and _da_-GAL4/UAS-dTSC2 (closed squares) flies were: on 0.1% yeast extract, _da_-Gal4/+, 33 days (n = 125) and _da_-GAL4/UAS-dTSC2, 30 days (n = 164); on 0.3% yeast extract, _da_-Gal4/+, 31 days (n = 139) and _da_-GAL4/UAS-dTSC2, 33 days (n = 138); on 1% yeast extract, _da_-Gal4/+, 30 days (n = 182) and _da_-GAL4/UAS-dTSC2, 34 days (n = 164); on 3% yeast extract, _da_-Gal4/+, 28 days (n = 181) and _da_-GAL4/UAS-dTSC2, 34 days (n = 154); on 9% yeast extract, _da_-Gal4/+, 12 days (n = 150) and _da_-GAL4/UAS-dTSC2, 24 days (n = 156). Mean lifespan differences between flies overexpressing dTsc2 and controls were −9% (p < 0.3), +5% (p < 0.03), +13% (p < 0.0001), +23% (p < 0.0001), and 95% (p < 0.0001) when lifespan was measured on fly food containing 0.1%, 0.3%, 1%, 3%, and 9% yeast extract, respectively.
Similar articles
- Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling.
Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Gao X, et al. Nat Cell Biol. 2002 Sep;4(9):699-704. doi: 10.1038/ncb847. Nat Cell Biol. 2002. PMID: 12172555 - Tuberous sclerosis complex tumor suppressor-mediated S6 kinase inhibition by phosphatidylinositide-3-OH kinase is mTOR independent.
Jaeschke A, Hartkamp J, Saitoh M, Roworth W, Nobukuni T, Hodges A, Sampson J, Thomas G, Lamb R. Jaeschke A, et al. J Cell Biol. 2002 Oct 28;159(2):217-24. doi: 10.1083/jcb.jcb.200206108. Epub 2002 Oct 28. J Cell Biol. 2002. PMID: 12403809 Free PMC article. - Rhebbing up mTOR: new insights on TSC1 and TSC2, and the pathogenesis of tuberous sclerosis.
Kwiatkowski DJ. Kwiatkowski DJ. Cancer Biol Ther. 2003 Sep-Oct;2(5):471-6. doi: 10.4161/cbt.2.5.446. Cancer Biol Ther. 2003. PMID: 14614311 Review. - Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling.
Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tee AR, et al. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13571-6. doi: 10.1073/pnas.202476899. Epub 2002 Sep 23. Proc Natl Acad Sci U S A. 2002. PMID: 12271141 Free PMC article. - Role of TOR signaling in aging and related biological processes in Drosophila melanogaster.
Katewa SD, Kapahi P. Katewa SD, et al. Exp Gerontol. 2011 May;46(5):382-90. doi: 10.1016/j.exger.2010.11.036. Epub 2010 Dec 2. Exp Gerontol. 2011. PMID: 21130151 Free PMC article. Review.
Cited by
- Modulated Expression of the Protein Kinase GSK3 in Motor and Dopaminergic Neurons Increases Female Lifespan in Drosophila melanogaster.
Trostnikov MV, Veselkina ER, Krementsova AV, Boldyrev SV, Roshina NV, Pasyukova EG. Trostnikov MV, et al. Front Genet. 2020 Jun 30;11:668. doi: 10.3389/fgene.2020.00668. eCollection 2020. Front Genet. 2020. PMID: 32695143 Free PMC article. - MicroRNA in Aging: From Discovery to Biology.
Jung HJ, Suh Y. Jung HJ, et al. Curr Genomics. 2012 Nov;13(7):548-57. doi: 10.2174/138920212803251436. Curr Genomics. 2012. PMID: 23633914 Free PMC article. - Answering the ultimate question "what is the proximal cause of aging?".
Blagosklonny MV. Blagosklonny MV. Aging (Albany NY). 2012 Dec;4(12):861-77. doi: 10.18632/aging.100525. Aging (Albany NY). 2012. PMID: 23425777 Free PMC article. Review. - Ovariectomy uncouples lifespan from metabolic health and reveals a sex-hormone-dependent role of hepatic mTORC2 in aging.
Arriola Apelo SI, Lin A, Brinkman JA, Meyer E, Morrison M, Tomasiewicz JL, Pumper CP, Baar EL, Richardson NE, Alotaibi M, Lamming DW. Arriola Apelo SI, et al. Elife. 2020 Jul 28;9:e56177. doi: 10.7554/eLife.56177. Elife. 2020. PMID: 32720643 Free PMC article. - Management of multicellular senescence and oxidative stress.
Haines DD, Juhasz B, Tosaki A. Haines DD, et al. J Cell Mol Med. 2013 Aug;17(8):936-57. doi: 10.1111/jcmm.12074. Epub 2013 Jun 22. J Cell Mol Med. 2013. PMID: 23789967 Free PMC article. Review.
References
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. - PubMed
- Partridge L, Gems D. Mechanisms of ageing: public or private? Nat. Rev. Genet. 2002;3:165–175. - PubMed
- Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat. Genet. 1996;13:25–34. - PubMed
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. - PubMed
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 2002;4:699–704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases