Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase - PubMed (original) (raw)
Cdc14p/FEAR pathway controls segregation of nucleolus in S. cerevisiae by facilitating condensin targeting to rDNA chromatin in anaphase
Bi-Dar Wang et al. Cell Cycle. 2004 Jul.
Abstract
The condensin complex is the chief molecular machine of mitotic chromosome condensation. Nucleolar concentration of condensin in mitosis was previously shown to correlate with proficiency of rDNA condensation and segregation. To uncover the mechanisms facilitating this targeting we conducted a screen for mutants that impair mitotic condensin congression to the nucleolus. Mutants in the cdc14, esp1 and cdc5 genes, which encode FEAR-network components, showed the most prominent defects in mitotic condensin localization. We established that Cdc14p activity released by the FEAR pathway was required for proper condensin-to-rDNA targeting in anaphase. The MEN pathway was dispensable for condensin-to-rDNA targeting, however MEN-mediated release of Cdc14p later in anaphase allowed for proper, albeit delayed, condensin targeting to rDNA and successful segregation of nucleolus in the slk19 FEAR mutant. Although condensin was physically dislodged from rDNA in the cdc14 mutant, it was properly assembled, phosphorylated and chromatin-bound, suggesting that condensin was mis-targeted but active. This study identifies a novel pathway promoting condensin targeting to a specific chromosomal address, the rDNA locus.
Figures
Figure 1
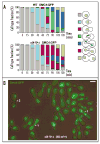
Condensin is delocalized in FEAR but not MEN mutants. (A) Smc4p-GFP signal is delocalized in FEAR mutants at 37°C. The esp1–1 strain (Table 1) was kept in mitosis with nocodazole. Scale bar 5 μm. (B) MEN mutants properly localize condensin to rDNA. Arrest conditions: cdc15-1 (37°C), _tem1_-Δ(3-hr shut-off of pGAL:NET1 at 23°C), _net1-_Δ (mitotic delay at 23°C). Strain genotypes in Table 1.
Figure 2
FEAR mutant slk19 is defective in anaphase concentration of condensin. (A) Condensin-to-nucleolus concentration in the synchronous cell cycle. Parallel analysis of 640-BY4741 (WT) and 640-4002451-BY4741 (_slk19_-Δ) strains released from alpha-factor arrest allowed comparative monitoring of Smc4p-GFP dynamics in the nucleus. Diffuse condensin, light green nucleus; nucleolus-targeted condensin, crescent-shaped dark-green subnuclear structure. (B) Micrograph of the 90-min point in the time-course analysis of the _slk19_-Δ strain. Arrow 1, unique class of cells undergoing anaphase with mistargeted condensin; Arrow 2, mid-to-late anaphase recapture of condensin by nucleolus. Scale bar 5 μm.
Figure 3
Diffuse condensin distribution in the cdc14 mutant correlates with a nucleolus segregation defect. (A) Micrograph and (B) quantification of relative distribution of Smc4p-GFP and Sik1p-mRFP in the 640-3AS426/SIK1:mRFP::kanMX cells after a temperature shift. Scale bar 5 μm. (C) Micrograph and (D) quantification of relative distribution of Smc4p-GFP and Sik1p-mRFP in the 640-1AS432/SIK1:mRFP::kanMX cells after a temperature shift. Scale bar 5 μm. (E) Same as in (C), with higher magnification.
Figure 4
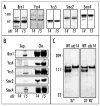
Mistargeted condensin is functional. (A) Western-blot of Brn1p-HA-precipitated condensin complex, showing comparison of condensin composition in cdc14 and cdc15 mutants at non-permissive temperature. Strains: Table 1. (B) Western-blot comparing chromatin-bound and soluble condensin fractions in cdc14 and cdc15 mutants at non-permissive temperature. Supernatant fractions (Sup.) are equivalent to 1/5 of the chromatin (Chr.) fractions. The strains are identical to (A). (C) Western-blot (anti-HA) analysis of Ycs5p-6xHis-3HA IMAC eluates from 3aAS453 (cdc14–1) and 6cAS453 (WT) incubated at two different conditions (See Materials and Methods). The upper Ycs5p band is a phosphorylated from. Mock purification (no 6xHis tag) produces no anti-HA-reactive bands at these stringent conditions (not shown).
Figure 5
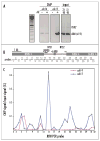
Condensin is physically de-localized from rDNA in the cdc14 cells. (A) Chromatin IP analysis of cdc14-1 BRN1:HA and cdc15-1 BRN1:HA strains arrested at 37°C for 2.5 hr. The RFB region, PCR probe #15 in (B), a known hot-spot of condensin localization to the rDNA repeat,, is used to assess the relative enrichment for condensin binding (Brn1p-HA ChIP). The TUB2 gene probe, which contains no binding sites for condensin (unpublished), is a negative control for duplex PCR. (B) Positions of PCR probes used for quantitative PCR analysis of ChIP in (C). PCR primer sequences were exactly as in reference . (C) Quantitative PCR analysis of Brn1p-HA ChIP from cdc14-1 and cdc15-1 strains arrested at 37°C for 2.5 hr. The quantitative PCR values from cycle 32 (see Materials and Methods) are plotted as ratios of precipitated DNA to total DNA (in %).
Figure 6
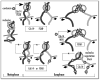
Interplay between Cdc14p, FEAR and MEN pathways in condensin localization to nucleolus in anaphase. Apparent chromosomal condensin distribution and corresponding success of rDNA/nucleolus segregation are shown for normal cell cycle, FEAR-deficient and Cdc14p deficient cells. Also shown is a supplementary role of MEN in condensin targeting when FEAR is inactivated.
Comment in
- Cdc14p regulates condensin binding to rDNA.
Strunnikov A. Strunnikov A. Cell Cycle. 2009 Apr 15;8(8):1114. doi: 10.4161/cc.8.8.8469. Epub 2009 Apr 15. Cell Cycle. 2009. PMID: 19305166 No abstract available.
Similar articles
- Cdc14 and condensin control the dissolution of cohesin-independent chromosome linkages at repeated DNA.
D'Amours D, Stegmeier F, Amon A. D'Amours D, et al. Cell. 2004 May 14;117(4):455-69. doi: 10.1016/s0092-8674(04)00413-1. Cell. 2004. PMID: 15137939 - Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase.
Sullivan M, Higuchi T, Katis VL, Uhlmann F. Sullivan M, et al. Cell. 2004 May 14;117(4):471-82. doi: 10.1016/s0092-8674(04)00415-5. Cell. 2004. PMID: 15137940 - Putting the brake on FEAR: Tof2 promotes the biphasic release of Cdc14 phosphatase during mitotic exit.
Waples WG, Chahwan C, Ciechonska M, Lavoie BD. Waples WG, et al. Mol Biol Cell. 2009 Jan;20(1):245-55. doi: 10.1091/mbc.e08-08-0879. Epub 2008 Oct 15. Mol Biol Cell. 2009. PMID: 18923139 Free PMC article. - A case of selfish nucleolar segregation.
Strunnikov AV. Strunnikov AV. Cell Cycle. 2005 Jan;4(1):113-7. doi: 10.4161/cc.4.1.1488. Epub 2005 Jan 21. Cell Cycle. 2005. PMID: 15655374 Review. - Cdc14 and the temporal coordination between mitotic exit and chromosome segregation.
Torres-Rosell J, Machín F, Aragón L. Torres-Rosell J, et al. Cell Cycle. 2005 Jan;4(1):109-12. doi: 10.4161/cc.4.1.1356. Epub 2005 Jan 10. Cell Cycle. 2005. PMID: 15611663 Review.
Cited by
- PICH promotes mitotic chromosome segregation: Identification of a novel role in rDNA disjunction.
Nielsen CF, Hickson ID. Nielsen CF, et al. Cell Cycle. 2016 Oct 17;15(20):2704-11. doi: 10.1080/15384101.2016.1222336. Epub 2016 Aug 26. Cell Cycle. 2016. PMID: 27565185 Free PMC article. - The Transient Inactivation of the Master Cell Cycle Phosphatase Cdc14 Causes Genomic Instability in Diploid Cells of Saccharomyces cerevisiae.
Quevedo O, Ramos-Pérez C, Petes TD, Machín F. Quevedo O, et al. Genetics. 2015 Jul;200(3):755-69. doi: 10.1534/genetics.115.177626. Epub 2015 May 12. Genetics. 2015. PMID: 25971663 Free PMC article. - Nutrient starvation promotes condensin loading to maintain rDNA stability.
Tsang CK, Li H, Zheng XS. Tsang CK, et al. EMBO J. 2007 Jan 24;26(2):448-58. doi: 10.1038/sj.emboj.7601488. Epub 2007 Jan 4. EMBO J. 2007. PMID: 17203076 Free PMC article. - The condensin complexes play distinct roles to ensure normal chromosome morphogenesis during meiotic division in Arabidopsis.
Smith SJ, Osman K, Franklin FC. Smith SJ, et al. Plant J. 2014 Oct;80(2):255-68. doi: 10.1111/tpj.12628. Epub 2014 Sep 4. Plant J. 2014. PMID: 25065716 Free PMC article. - Essential global role of CDC14 in DNA synthesis revealed by chromosome underreplication unrecognized by checkpoints in cdc14 mutants.
Dulev S, de Renty C, Mehta R, Minkov I, Schwob E, Strunnikov A. Dulev S, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14466-71. doi: 10.1073/pnas.0900190106. Epub 2009 Aug 7. Proc Natl Acad Sci U S A. 2009. PMID: 19666479 Free PMC article.
References
- Swedlow JR, Hirano T. The making of the mitotic chromosome: modern insights into classical questions. Mol Cell. 2003;11:557–69. - PubMed
- Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell. 2003;5:323–36. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases